7FHK
 
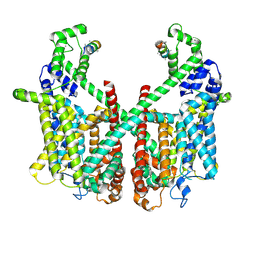 | | Structure of AtTPC1 with 1 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7F9L
 
 | | Crystal structure of the variable region of Plasmodium RIFIN #6 (PF3D7_1400600) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9M
 
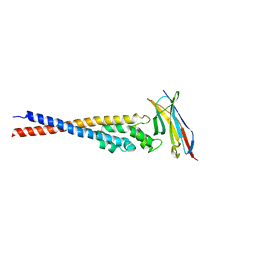 | | Crystal structure of the variable region of Plasmodium RIFIN #4 (PF3D7_1000500) in complex with LAIR1 (with T67L, N69S and A77T mutations) | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1, Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7F9K
 
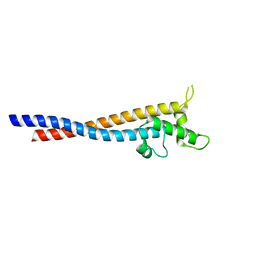 | | Crystal structure of the variable region of Plasmodium RIFIN #6(PF3D7_1400600) | | Descriptor: | Rifin | | Authors: | Xie, Y, Song, H, Li, X, Qi, J, Gao, G.F. | | Deposit date: | 2021-07-04 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of malarial parasite RIFIN-mediated immune escape against LAIR1.
Cell Rep, 36, 2021
|
|
7DLY
 
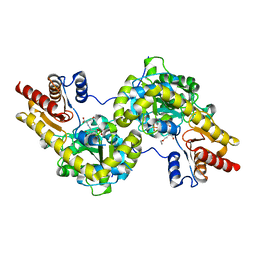 | | Crystal structure of Arabidopsis ACS7 mutant in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7 | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7DLW
 
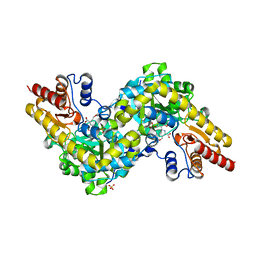 | | Crystal structure of Arabidopsis ACS7 in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7, SULFATE ION | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7F0X
 
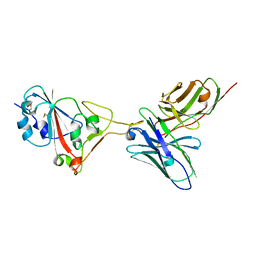 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F12
 
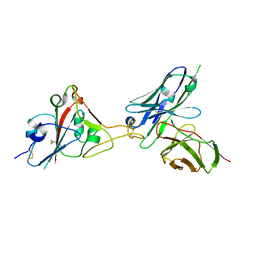 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
7F15
 
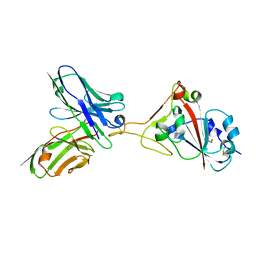 | | A SARS-CoV-2 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Spike protein S1 | | Authors: | Zhang, G, Li, X, Guo, Y, Wang, Y, Yuan, S. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A SARS-CoV-2 neutralizing
antibody
To Be Published
|
|
6KUZ
 
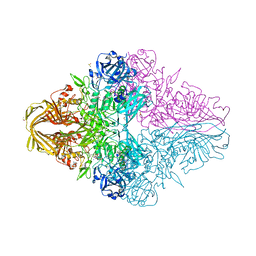 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSL01 | | Descriptor: | 3-(1,3-benzothiazol-2-yl)-2-[[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]methoxy]-5-methyl-benzaldehyde, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Li, X.K, Guo, Y, Li, J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | First-generation species-selective chemical probes for fluorescence imaging of human senescence-associated beta-galactosidase.
Chem Sci, 11, 2020
|
|
5VK0
 
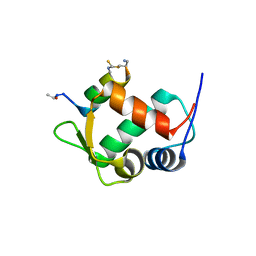 | |
5VK1
 
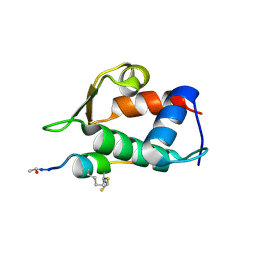 | |
8K82
 
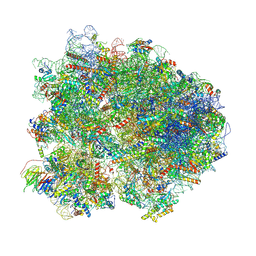 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, Not5 and P-site tRNA | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-28 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8JW0
 
 | | PSI-AcpPCI supercomplex from Amphidinium carterae | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JZF
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, X.Y, Li, Z.H, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JZE
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K2D
 
 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
5UR1
 
 | | FGFR1 kinase domain complex with SN37333 in reversible binding mode | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{1-[4-(dimethylamino)but-2-enoyl]piperidin-4-yl}-7-(phenylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1 | | Authors: | Yosaatmadja, Y, Paik, W.-K, Smaill, J.B, Squire, C.J. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Oxo-3, 4-dihydropyrimido[4, 5-d]pyrimidinyl derivatives as new irreversible pan fibroblast growth factor receptor (FGFR) inhibitors.
Eur J Med Chem, 135, 2017
|
|
2ESW
 
 | |
4LEZ
 
 | | Structure of mouse cGAS bound to an 18bp DNA and cGAS product | | Descriptor: | 18bp dsDNA, Cyclic GMP-AMP synthase, ZINC ION, ... | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
4LEV
 
 | | Structure of human cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
4LEW
 
 | | Structure of human cGAS | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2014-01-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
4LEY
 
 | | Structure of mouse cGAS bound to 18 bp DNA | | Descriptor: | 18 bp dsDNA, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Li, P. | | Deposit date: | 2013-06-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization.
Immunity, 39, 2013
|
|
1BBA
 
 | |
5YYF
 
 | |
