2C7C
 
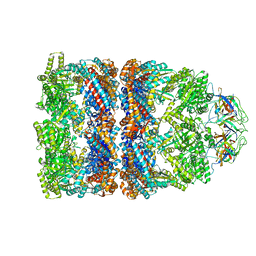 | | FITTED COORDINATES FOR GROEL-ATP7-GROES CRYO-EM COMPLEX (EMD-1180) | | Descriptor: | 10 KDA CHAPERONIN MOLECULE: GROES, PROTEIN CPN10, GROES PROTEIN, ... | | Authors: | Ranson, N.A, Clare, D.K, Farr, G.W, Houldershaw, D, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2005-11-22 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Allosteric Signalling of ATP Hydrolysis in Groel-Groes Complexes.
Nat.Struct.Mol.Biol., 13, 2006
|
|
7LXW
 
 | |
2CHE
 
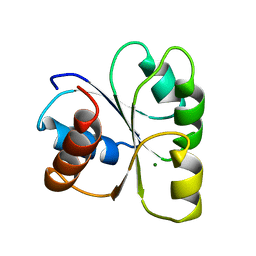 | | STRUCTURE OF THE MG2+-BOUND FORM OF CHEY AND MECHANISM OF PHOSPHORYL TRANSFER IN BACTERIAL CHEMOTAXIS | | Descriptor: | CHEY, MAGNESIUM ION | | Authors: | Stock, A, Martinez-Hackert, E, Rasmussen, B, West, A, Stock, J, Ringe, D, Petsko, G. | | Deposit date: | 1994-01-17 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis.
Biochemistry, 32, 1993
|
|
7LY0
 
 | |
2CKM
 
 | | Torpedo californica acetylcholinesterase complexed with alkylene- linked bis-tacrine dimer (7 carbon linker) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, N,N'-DI-1,2,3,4-TETRAHYDROACRIDIN-9-YLHEPTANE-1,7-DIAMINE | | Authors: | Brumshtein, B, Rydberg, E.H, Greenblatt, H.M, Wong, D.M, Shaya, D, Williams, L.D, Carlier, P.R, Pang, Y.P, Silman, I, Sussman, J.L. | | Deposit date: | 2006-04-20 | | Release date: | 2006-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Complexes of alkylene-linked tacrine dimers with Torpedo californica acetylcholinesterase: Binding of Bis5-tacrine produces a dramatic rearrangement in the active-site gorge.
J. Med. Chem., 49, 2006
|
|
7LXZ
 
 | |
8YK5
 
 | |
7LY2
 
 | |
8XFY
 
 | | The Crystal Structure of RSK2 from Biortus. | | Descriptor: | 2,6-bis(fluoranyl)-4-[4-(4-morpholin-4-ylphenyl)pyridin-3-yl]phenol, Ribosomal protein S6 kinase alpha-3 | | Authors: | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | Deposit date: | 2023-12-14 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of RSK2 from Biortus.
To Be Published
|
|
7LXX
 
 | |
7LXY
 
 | |
3BPQ
 
 | |
7LY3
 
 | |
3BRW
 
 | | Structure of the Rap-RapGAP complex | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Scrima, A, Thomas, C, Deaconescu, D, Wittinghofer, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues
Embo J., 27, 2008
|
|
3BU2
 
 | | Crystal structure of a tRNA-binding protein from Staphylococcus saprophyticus subsp. saprophyticus. Northeast Structural Genomics Consortium target SyR77 | | Descriptor: | Putative tRNA-binding protein | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-31 | | Release date: | 2008-01-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a tRNA-binding protein from Staphylococcus saprophyticus subsp. saprophyticus.
To be Published
|
|
2CIE
 
 | | Complexes of Dodecin with Flavin and Flavin-like Ligands | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Grininger, M, Seiler, F, Zeth, K, Oesterhelt, D. | | Deposit date: | 2006-03-20 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dodecin Sequesters Fad in Closed Conformation from the Aqueous Solution.
J.Mol.Biol., 364, 2006
|
|
8XKF
 
 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
2X99
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with NADPH | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Boumis, G, Brunori, M, Miele, A.E, Saccoccia, F, Bellelli, A. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping the Catalytic Cycle of Schistosoma Mansoni Thioredoxin Glutathione Reductase by X-Ray Crystallography
J.Biol.Chem., 285, 2010
|
|
3BVQ
 
 | | Crystal Structure of Apo NotI Restriction Endonuclease | | Descriptor: | FE (III) ION, NotI restriction endonuclease, SULFATE ION | | Authors: | Lambert, A.R, Sussman, D, Shen, B, Stoddard, B.L. | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-22 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Rare-Cutting Restriction Endonuclease NotI Reveal a Unique Metal Binding Fold Involved in DNA Binding.
Structure, 16, 2008
|
|
3BW6
 
 | | Crystal structure of the longin domain of yeast Ykt6 | | Descriptor: | SULFATE ION, Synaptobrevin homolog YKT6 | | Authors: | Pylypenko, O, Schonichen, A, Ludwig, D, Ungermann, C, Goody, R.S, Rak, A, Geyer, M. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Farnesylation of the SNARE protein Ykt6 increases its stability and helical folding.
J.Mol.Biol., 377, 2008
|
|
3BZC
 
 | |
1PRE
 
 | | PROAEROLYSIN | | Descriptor: | PROAEROLYSIN | | Authors: | Parker, M.W, Buckley, J.T, Postma, J.P.M, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1995-09-15 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states.
Nature, 367, 1994
|
|
2CCB
 
 | | Complexes of Dodecin with Flavin and Flavin-like Ligands | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RIBOFLAVIN, ... | | Authors: | Grininger, M, Zeth, K, Oesterhelt, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dodecins: A Family of Lumichrome Binding Proteins.
J.Mol.Biol., 357, 2006
|
|
2CCR
 
 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, TRIETHYLENE GLYCOL, YVFO, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-03-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
2CIF
 
 | | Complexes of Dodecin with Flavin and Flavin-like Ligands | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Grininger, M, Seiler, F, Zeth, K, Oesterhelt, D. | | Deposit date: | 2006-03-20 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dodecin Sequesters Fad in Closed Conformation from the Aqueous Solution.
J.Mol.Biol., 364, 2006
|
|
