4D3E
 
 | | Tetramer of IpaD, modified from 2J0O, fitted into negative stain electron microscopy reconstruction of the wild type tip complex from the type III secretion system of Shigella flexneri | | Descriptor: | INVASIN IPAD | | Authors: | Cheung, M, Shen, D.-K, Makino, F, Kato, T, Roehrich, D, Martinez-Argudo, I, Walker, M.L, Murillo, I, Liu, X, Pain, M, Brown, J, Frazer, G, Mantell, J, Mina, P, Todd, T, Sessions, R.B, Namba, K, Blocker, A.J. | | Deposit date: | 2014-10-21 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Three-Dimensional Electron Microscopy Reconstruction and Cysteine-Mediated Crosslinking Provide a Model of the T3Ss Needle Tip Complex.
Mol.Microbiol., 95, 2015
|
|
7XHQ
 
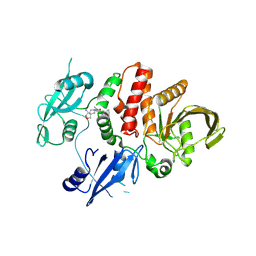 | | Small-molecule Allosteric Regulation Mechanism of SHP2 | | Descriptor: | 2-[(3S,4R)-4-azanyl-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl]-5-[2,3-bis(chloranyl)phenyl]-3-methyl-pyrrolo[2,1-f][1,2,4]triazin-4-one, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, Y, Zhu, J, Yu, K, Liu, B. | | Deposit date: | 2022-04-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the SHP2 allosteric inhibitor 2-((3R,4R)-4-amino-3-methyl-2-oxa-8-azaspiro[4.5]decan-8-yl)-5-(2,3-dichlorophenyl)-3-methylpyrrolo[2,1-f][1,2,4] triazin-4(3H)-one.
J Enzyme Inhib Med Chem, 38, 2023
|
|
6P4H
 
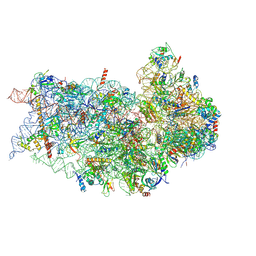 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, IAPV-IRES, RACK1, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6V9S
 
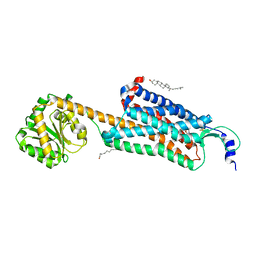 | | Structure-based development of subtype-selective orexin 1 receptor antagonists | | Descriptor: | CHOLESTEROL, OLEIC ACID, Orexin receptor type 1,GlgA glycogen synthase chimera, ... | | Authors: | Hellmann, J, Drabek, M, Yin, J, Huebner, H, Kraus, F, Proell, T, Weikert, D, Kolb, P, Rosenbaum, D.M, Gmeiner, P. | | Deposit date: | 2019-12-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-based development of a subtype-selective orexin 1 receptor antagonist.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5D7X
 
 | |
6PLX
 
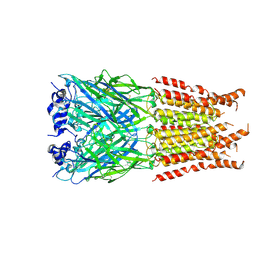 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, desensitized state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PMO
 
 | |
7Y5E
 
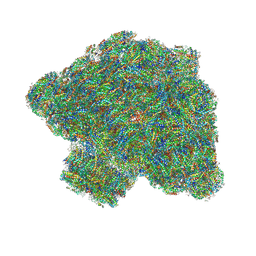 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
6PNY
 
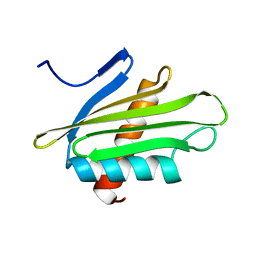 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
7XKZ
 
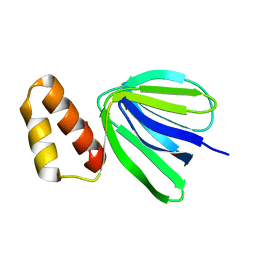 | | Solution structure of subunit epsilon of the Mycobacterium abscessus F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Grueber, G, Harikishore, A, Wong, C.F, Prya, R, Dick, T. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Atomic solution structure of Mycobacterium abscessus F-ATP synthase subunit epsilon and identification of Ep1MabF1 as a targeted inhibitor.
Febs J., 289, 2022
|
|
6P5I
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P4G
 
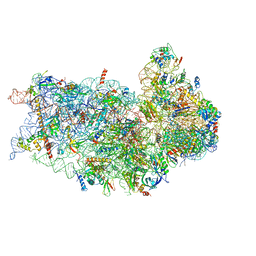 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | Descriptor: | 18S rRNA, IAPV-IRES, RACK1, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-27 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
4TWE
 
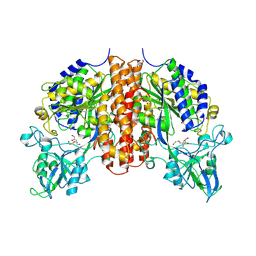 | | Structure of ligand-free N-acetylated-alpha-linked-acidic-dipeptidase like protein (NAALADaseL) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Barinka, C, Lubkowski, J, Sacha, P, Konvalinka, J. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of a novel aminopeptidase from human intestine.
J.Biol.Chem., 290, 2015
|
|
5DF6
 
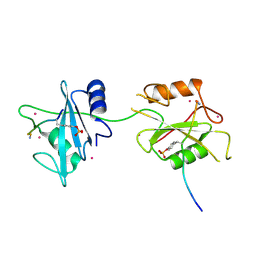 | | Crystal structure of PTPN11 tandem SH2 domains in complex with a TXNIP peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, UNKNOWN ATOM OR ION, txnip | | Authors: | Dong, A, Li, W, Tempel, W, Liu, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
2G6L
 
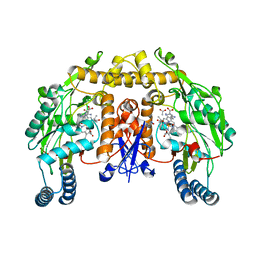 | | Structure of rat nNOS heme domain (BH2 bound) complexed with NO | | Descriptor: | 7,8-DIHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Li, H, Igarashi, J, Jamal, J, Yang, W, Poulos, T.L. | | Deposit date: | 2006-02-24 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural studies of constitutive nitric oxide synthases with diatomic ligands bound.
J.Biol.Inorg.Chem., 11, 2006
|
|
4L3B
 
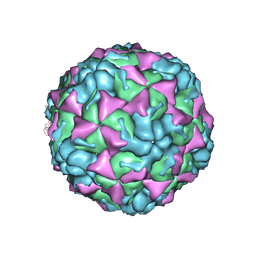 | | X-ray structure of the HRV2 A particle uncoating intermediate | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Vives-Adrian, L, Querol-Audi, J, Garriga, D, Pous, J, Verdaguer, N. | | Deposit date: | 2013-06-05 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Uncoating of common cold virus is preceded by RNA switching as determined by X-ray and cryo-EM analyses of the subviral A-particle.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2G74
 
 | |
9F9Y
 
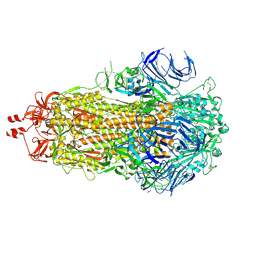 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
2FN4
 
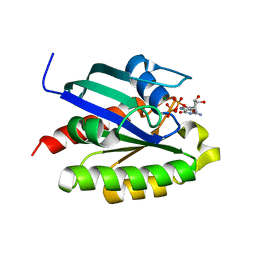 | | The crystal structure of human Ras-related protein, RRAS, in the GDP-bound state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein R-Ras | | Authors: | Turnbull, A.P, Elkins, J.M, Gileadi, C, Burgess, N, Salah, E, Papagrigoriou, E, Debreczeni, J, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-10 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of human Ras-related protein, RRAS, in the GDP-bound state
To be Published
|
|
5CXC
 
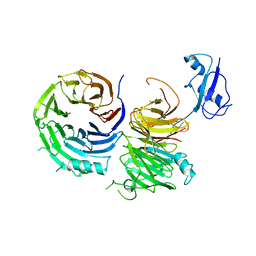 | | Structure of Ytm1 bound to the C-terminal domain of Erb1 in P 65 2 2 space group | | Descriptor: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | Authors: | Wegrecki, M, Bravo, J. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
5CYK
 
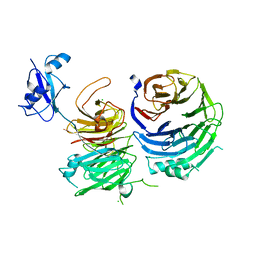 | | Structure of Ytm1 bound to the C-terminal domain of Erb1-R486E | | Descriptor: | CHLORIDE ION, Ribosome biogenesis protein ERB1, Ribosome biogenesis protein YTM1 | | Authors: | Wegrecki, M, Bravo, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of Erb1-Ytm1 complex reveals the functional importance of a high-affinity binding between two beta-propellers during the assembly of large ribosomal subunits in eukaryotes.
Nucleic Acids Res., 43, 2015
|
|
6CDX
 
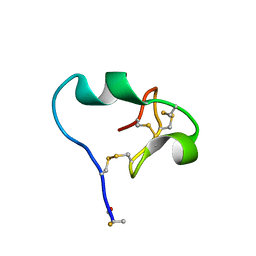 | | High-resolution crystal structure of fluoropropylated cystine knot, binding to alpha-5 beta-6 integrin | | Descriptor: | cystine knot (fluoropropylated) | | Authors: | Kimura, R, Nix, J, Bongura, C, Chakraborti, S, Gambhir, S, Filipp, F.V. | | Deposit date: | 2018-02-09 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Evaluation of integrin alpha v beta6cystine knot PET tracers to detect cancer and idiopathic pulmonary fibrosis.
Nat Commun, 10, 2019
|
|
4CSD
 
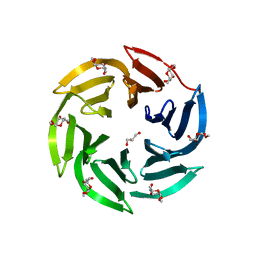 | | Structure of Monomeric Ralstonia solanacearum lectin | | Descriptor: | FUCOSE-BINDING LECTIN PROTEIN, GLYCEROL, methyl alpha-L-fucopyranoside | | Authors: | Arnaud, J, Trundle, K, Claudinon, J, Audfray, A, Varrot, A, Romer, W, Imberty, A. | | Deposit date: | 2014-03-06 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Membrane Deformation by Neolectins with Engineered Glycolipid Binding Sites.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6PIV
 
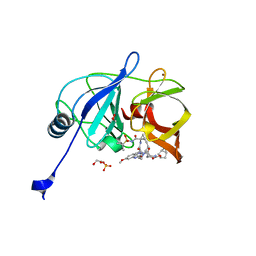 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
5D2U
 
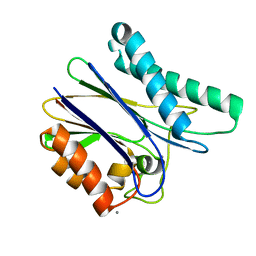 | | Crystal structure of tPphA Variant - H39A | | Descriptor: | CALCIUM ION, Protein serin-threonin phosphatase | | Authors: | Su, J. | | Deposit date: | 2015-08-06 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of a Cyanobacterial PP2C Phosphatase Reveals Insights into Catalytic Mechanism and Substrate Recognition
Catalysts, 2016
|
|
