7GDN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-0e996074-1 (Mpro-x11159) | | Descriptor: | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDY
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with LOR-NOR-30067bb9-6 (Mpro-x11258) | | Descriptor: | (3S)-5-bromo-1-[(2-ethoxyphenyl)methyl]-3-hydroxy-1,3-dihydro-2H-indol-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDC
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-0a73fcb8-7 (Mpro-x10942) | | Descriptor: | (4R)-6-chloro-N-[4-(hydroxymethyl)pyridin-3-yl]-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
8S4H
 
 | |
8S4K
 
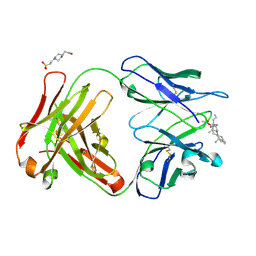 | | Crystal structure of Fab-2B1 in complex with rocuronium | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab-2B1 VH, Fab-2B1 VL, ... | | Authors: | Saul, F.A, Haouz, A, Bruhns, P. | | Deposit date: | 2024-02-21 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rocuronium-specific antibodies drive perioperative anaphylaxis but can also function as reversal agents in preclinical models.
Sci Transl Med, 16, 2024
|
|
1TU2
 
 | | THE COMPLEX OF NOSTOC CYTOCHROME F AND PLASTOCYANIN DETERMIN WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | Descriptor: | Apocytochrome f, COPPER (II) ION, HEME C, ... | | Authors: | Diaz-Moreno, I, Diaz-Quintana, A, De la Rosa, M.A, Ubbink, M. | | Deposit date: | 2004-06-24 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex between plastocyanin and cytochrome f from the cyanobacterium Nostoc sp. PCC 7119 as determined by paramagnetic NMR. The balance between electrostatic and hydrophobic interactions within the transient complex determines the relative orientation of the two proteins.
J.Biol.Chem., 280, 2005
|
|
8OE6
 
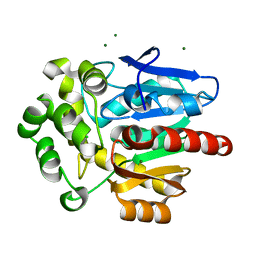 | | Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
8OE2
 
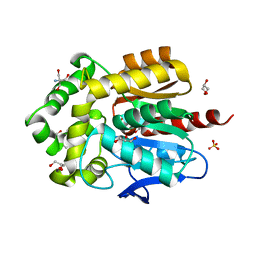 | | Structure of hyperstable haloalkane dehalogenase variant DhaA223 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
1DMF
 
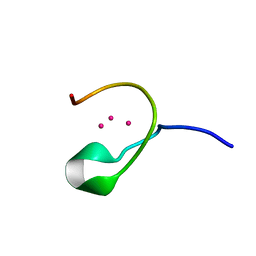 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DME
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DMD
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DMC
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
4Z6K
 
 | |
6KHT
 
 | | Chimeric beta-glucosidase Cel1b-H13 | | Descriptor: | Glycoside hydrolase family 1 | | Authors: | Niu, K.L. | | Deposit date: | 2019-07-16 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.305 Å) | | Cite: | Chimeric beta-glucosidase Cel1b-H13
To Be Published
|
|
6S6E
 
 | |
3NZ0
 
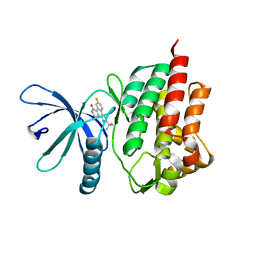 | | Non-phosphorylated TYK2 kinase with CMP6 | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new regulatory switch in a JAK protein kinase.
Proteins, 79, 2011
|
|
3NYX
 
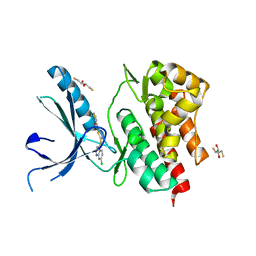 | | Non-phosphorylated TYK2 JH1 domain with Quinoline-Thiadiazole-Thiophene Inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, N-{5-[(7-chloroquinolin-4-yl)sulfanyl]-1,3,4-thiadiazol-2-yl}thiophene-2-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new regulatory switch in a JAK protein kinase.
Proteins, 79, 2011
|
|
6S97
 
 | |
3N2J
 
 | | Azurin H117G, oxidized form | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Hoffmann, M, Alagaratnam, S, Canters, G.W, Einsle, O. | | Deposit date: | 2010-05-18 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Probing the reactivity of different forms of azurin by flavin photoreduction.
Febs J., 278, 2011
|
|
6XT8
 
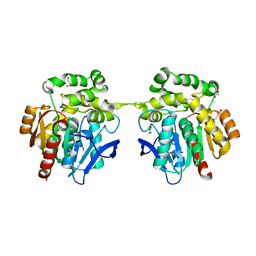 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Markova, K, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
6XTC
 
 | |
3PZE
 
 | | JNK1 in complex with inhibitor | | Descriptor: | 3-(carbamoylamino)-5-phenylthiophene-2-carboxamide, Mitogen-activated protein kinase 8, SULFATE ION | | Authors: | Xue, Y. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(piperidin-3-yl)-3-ureidothiophene-2-carboxamide (AZD7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
6YN2
 
 | |
6TY7
 
 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
5NQA
 
 | | Crystal structure of GalNAc-T4 in complex with the monoglycopeptide 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, GLYCEROL, ... | | Authors: | de las Rivas, M, Lira-Navarrete, E, Daniel, E.J.P, Companon, I, Coelho, H, Diniz, A, Jimenez-Barbero, J, Peregrina, J.M, Clausen, H, Corzana, F, Marcelo, F, Jimenez-Oses, G, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences.
Nat Commun, 8, 2017
|
|
