6IMH
 
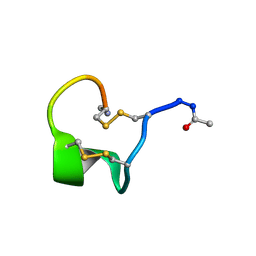 | | Solution Structure of Bicyclic Peptide pb-18 | | Descriptor: | (ACE)-GLY-CYS-PRO-CYS-GLU-PRO-SER-TYR-LEU-CYS-PRO-TRP-LEU-PRO-GLY-CYS-(NH2) | | Authors: | Yao, H, Lin, P, Zha, J, Zha, M, Zhao, Y, Wu, C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Ordered and Isomerically Stable Bicyclic Peptide Scaffolds Constrained through Cystine Bridges and Proline Turns.
Chembiochem, 20, 2019
|
|
6ISK
 
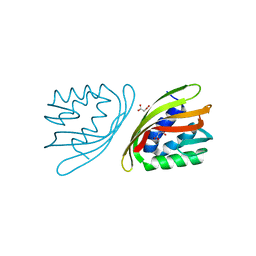 | | SnoaL-like cyclase XimE | | Descriptor: | 1,2-ETHANEDIOL, MALONIC ACID, XimE, ... | | Authors: | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
6IMG
 
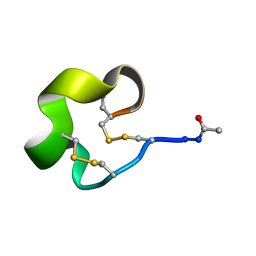 | | Solution Structure of Bicyclic Peptide pb-13 | | Descriptor: | (ACE)-GLY-CYS-PRO-CYS-ILE-TRP-PRO-GLU-LEU-CYS-PRO-TRP-ILE-ARG-SER-CYS-(NH2) | | Authors: | Yao, H, Lin, P, Zha, J, Zha, M, Zhao, Y, Wu, C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Ordered and Isomerically Stable Bicyclic Peptide Scaffolds Constrained through Cystine Bridges and Proline Turns.
Chembiochem, 20, 2019
|
|
6I2K
 
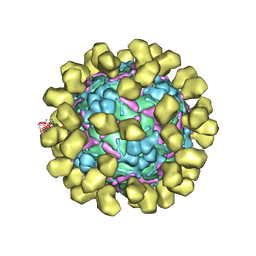 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
6JAK
 
 | | OtsA apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-24 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
6JBI
 
 | | Structure of Tps1 apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Yi, L, Wang, D, Liu, J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
5H3V
 
 | | Crystal structure of a Type IV Secretion System Component CagX in Helicobacter pylori | | Descriptor: | Cag8, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Zhang, J, Wu, Y, Zhao, Y, Sun, L, Keegan, R.M, Liu, Y, Isupov, M.N. | | Deposit date: | 2016-10-27 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the type IV secretion system component CagX from Helicobacter pylori
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5F54
 
 | | Structure of RecJ complexed with dTMP | | Descriptor: | MANGANESE (II) ION, Single-stranded-DNA-specific exonuclease, THYMIDINE-5'-PHOSPHATE | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5F55
 
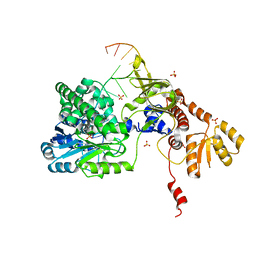 | | Structure of RecJ complexed with DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*C)-3'), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5GHY
 
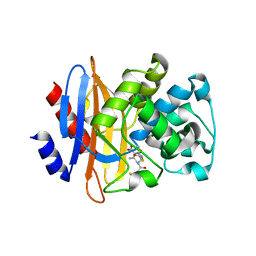 | |
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
8H1D
 
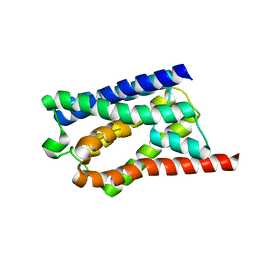 | | Solid-state NMR Structure of Aquaporin Z in its Native Cellular Membranes | | Descriptor: | Aquaporin Z | | Authors: | Xie, H, Zhao, Y, Zhao, W, Chen, Y, Liu, M, Yang, J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure determination of a membrane protein in E. coli cellular inner membrane.
Sci Adv, 9, 2023
|
|
7ND3
 
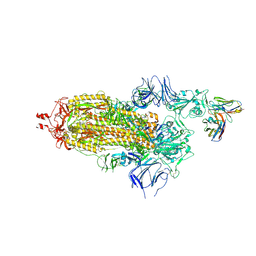 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-40 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND4
 
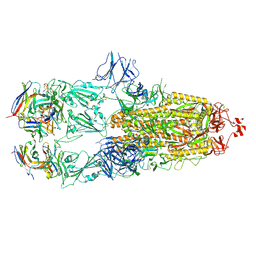 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-88 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-88 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDC
 
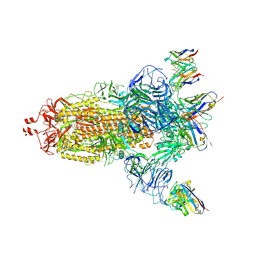 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND7
 
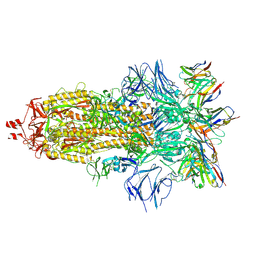 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND9
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-253H55L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDA
 
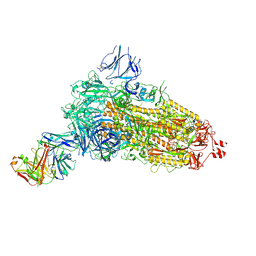 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-253H55L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND5
 
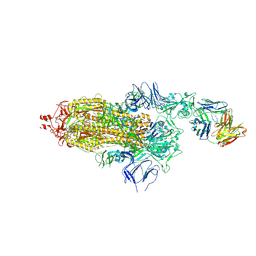 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDD
 
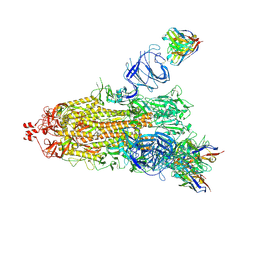 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-159 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND6
 
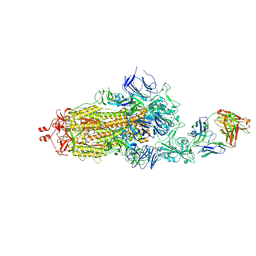 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDB
 
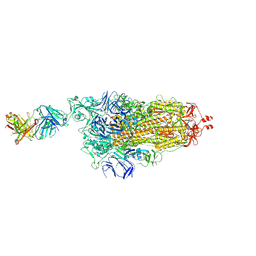 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-253H165L Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND8
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-384 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-384 Fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2021-01-30 | | Release date: | 2021-03-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
6OCP
 
 | | Crystal structure of a human GABAB receptor peptide bound to KCTD16 T1 | | Descriptor: | BTB/POZ domain-containing protein KCTD16, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8F97
 
 | | Compound 5 bound to procaspase-6 | | Descriptor: | 1,2-ETHANEDIOL, 2-({[(2M)-[2,3'-bipyridin]-2'-yl]amino}methyl)-5-fluorophenol, CHLORIDE ION, ... | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
