6PDI
 
 | | Human PIM1 bound to benzothiophene inhibitor 224 | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-{[(3-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, GLYCEROL, ... | | Authors: | Godoi, P.H.C, Fala, A.M, Santiago, A.S, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human PIM1
To Be Published
|
|
6PDP
 
 | | Human PIM1 bound to benzothiophene inhibitor 379 | | Descriptor: | 5-[2-(acetylamino)-1-benzothiophen-4-yl]-N-cyclopropylthiophene-2-carboxamide, Peptide, SULFATE ION, ... | | Authors: | Godoi, P.H.C, Sriranganadane, D, Santiago, A.S, Fala, A.M, Ramos, P.Z, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human PIM1
To Be Published
|
|
8CPH
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 (inactive form) | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPI
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPJ
 
 | | Crystal structure of PPAR gamma (PPARG) in an inactive form | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8G43
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(methylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(methylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
8G44
 
 | | Structure of HDAC6 zinc-finger ubiquitin binding domain in complex with 3-(3-(2-(benzylamino)-2-oxoethyl)-4-oxo-3,4-dihydroquinazolin-2-yl)propanoic acid | | Descriptor: | 3-{3-[2-(benzylamino)-2-oxoethyl]-4-oxo-3,4-dihydroquinazolin-2-yl}propanoic acid, Histone deacetylase 6, ZINC ION | | Authors: | Harding, R.J, Franzoni, I, Mann, M.K, Szewczyk, M, Mirabi, B, Owens, D.D.G, Ackloo, S, Scheremetjew, A, Juarez-Ornelas, K.A, Sanichar, R, Baker, R.J, Dank, C, Brown, P.J, Barsyte-Lovejoy, D, Santhakumar, V, Schapira, M, Lautens, M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Characterization of a Chemical Probe Targeting the Zinc-Finger Ubiquitin-Binding Domain of HDAC6.
J.Med.Chem., 66, 2023
|
|
6HPZ
 
 | | Crystal structure of ENL (MLLT1) in complex with acetyllysine | | Descriptor: | 1,2-ETHANEDIOL, N(6)-ACETYLLYSINE, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
5WP3
 
 | | Crystal Structure of EED in complex with EB22 | | Descriptor: | EB22, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Zhu, L, Moody, J.D, Baker, D, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First critical repressive H3K27me3 marks in embryonic stem cells identified using designed protein inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6HPW
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 20 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HQ0
 
 | | Crystal structure of ENL (MLLT1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6RU8
 
 | | Crystal structure of Casein Kinase I delta (CK1d) in complex with triple phosphorylated p63 PAD3P peptide | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Tuppi, M, Gebel, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Dotsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | p63 uses a switch-like mechanism to set the threshold for induction of apoptosis.
Nat.Chem.Biol., 16, 2020
|
|
6HPX
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 19 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[(3-chlorophenyl)methyl]-1-(2-pyrrolidin-1-ylethyl)benzimidazole-5-carboxamide | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HPY
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 12 | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]propanoic acid, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
6HRH
 
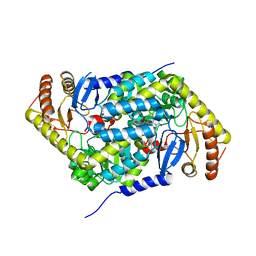 | | Structure of human erythroid-specific 5'-aminolevulinate synthase, ALAS2 | | Descriptor: | 5-aminolevulinate synthase, erythroid-specific, mitochondrial, ... | | Authors: | Bailey, H.J, Shrestha, L, Rembeza, E, Newman, J, Kupinska, K, Diaz-saez, L, Kennedy, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-27 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human erythroid-specific 5'-aminolevulinate synthase, ALAS2
To Be Published
|
|
9EPW
 
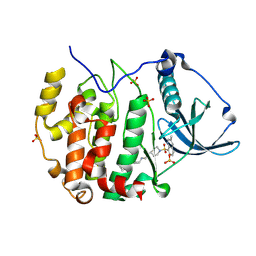 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3336 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[(3-chloranyl-4-phenyl-phenyl)methyl]-2-[1-(3-methylquinolin-8-yl)sulfonylpiperidin-4-yl]ethanamine | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPZ
 
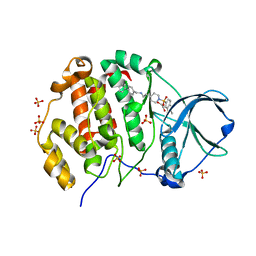 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3337 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EQ1
 
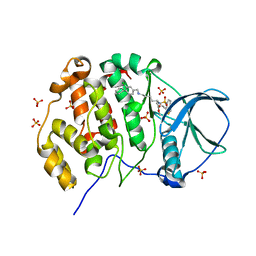 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJM24 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Moeckel, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
3KVW
 
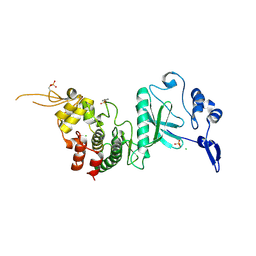 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | Descriptor: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
9EQ0
 
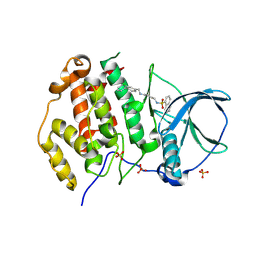 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGJG12 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[4-[(3-chloranyl-4-phenyl-phenyl)methylamino]butyl]isoquinoline-5-sulfonamide | | Authors: | Kraemer, A, Greco, F, Gerninghaus, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPV
 
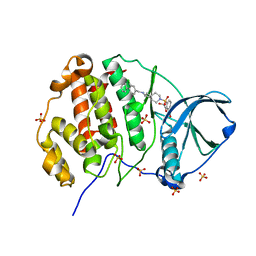 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC333 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[2-[(3-chloranyl-4-phenyl-phenyl)methylamino]-7-azaspiro[3.5]nonan-7-yl]sulfonyl]-1,3-dimethyl-benzimidazol-2-one, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
9EPY
 
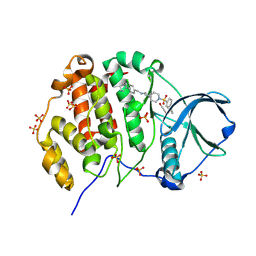 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with allosteric ligand FGC3330 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Kraemer, A, Greco, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Synthesis and evaluation of chemical linchpins for highly selective CK2 alpha targeting.
Eur.J.Med.Chem., 276, 2024
|
|
3KUQ
 
 | | Crystal structure of the DLC1 RhoGAP domain | | Descriptor: | Rho GTPase-activating protein 7, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, MacKenzie, F, Shen, L, Zhong, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DLC1 RhoGAP domain
to be published
|
|
6I5I
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6I5K
 
 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN345 (derivative of compound 12h) | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-(3-propan-2-yloxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
