1JY0
 
 | |
6A4V
 
 | | Open Reading frame 49 | | Descriptor: | 49 protein | | Authors: | Hwang, K.Y, Song, M.J, Kim, J.S, Cheong, W.C. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based mechanism of action of a viral poly(ADP-ribose) polymerase 1-interacting protein facilitating virus replication.
Iucrj, 5, 2018
|
|
1NS4
 
 | |
1NSX
 
 | |
1NSR
 
 | |
1NSM
 
 | |
1NSZ
 
 | |
1NSV
 
 | |
1NS8
 
 | |
1NSU
 
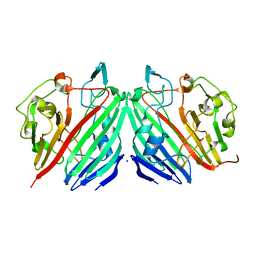 | |
6KBO
 
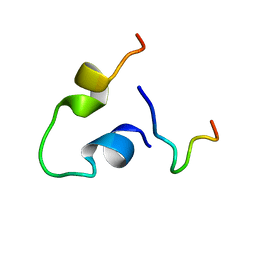 | | Three-dimensional LPS bound structure of VG16KRKP-KYE28. | | Descriptor: | Heparin cofactor 2, VG16KRKP | | Authors: | Ilyas, H, Bhunia, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism.
J.Biol.Chem., 294, 2019
|
|
1NS2
 
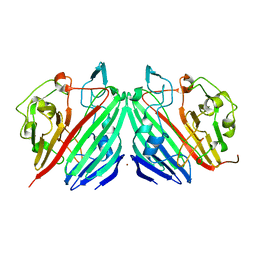 | |
1NSS
 
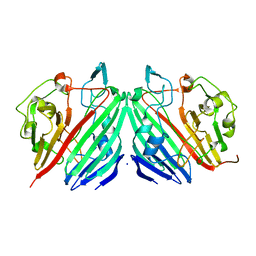 | |
1NS0
 
 | |
1NS7
 
 | |
6KBV
 
 | |
7ERQ
 
 | |
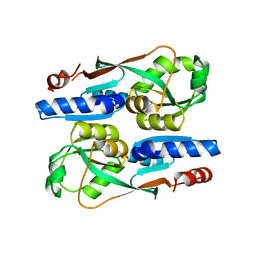
7ERP
 
 | |
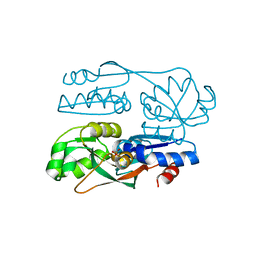
7FDF
 
 | |
1W7P
 
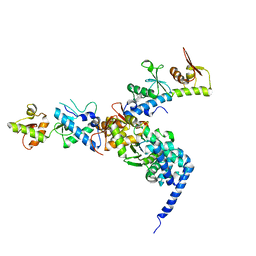 | | The crystal structure of endosomal complex ESCRT-II (VPS22/VPS25/VPS36) | | Descriptor: | VPS22, YPL002C, VPS25, ... | | Authors: | Teo, H, Perisic, O, Gonzalez, B, Williams, R.L. | | Deposit date: | 2004-09-07 | | Release date: | 2004-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Escrt-II, an Endosome-Associated Complex Required for Protein Sorting: Crystal Structure and Interactions with Escrt-III and Membranes
Dev.Cell, 7, 2004
|
|
1AO7
 
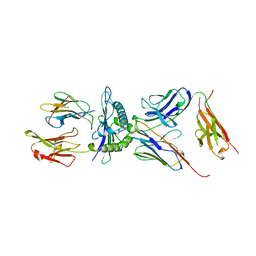 | | COMPLEX BETWEEN HUMAN T-CELL RECEPTOR, VIRAL PEPTIDE (TAX), AND HLA-A 0201 | | Descriptor: | BETA-2 MICROGLOBULIN, ETHYL MERCURY ION, HLA-A 0201, ... | | Authors: | Garboczi, D.N, Ghosh, P, Utz, U, Fan, Q.R, Biddison, W.E, Wiley, D.C. | | Deposit date: | 1997-07-21 | | Release date: | 1997-09-17 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the complex between human T-cell receptor, viral peptide and HLA-A2.
Nature, 384, 1996
|
|
6BFA
 
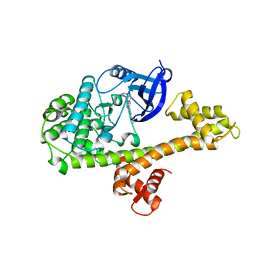 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with inhibitor UW1553 | | Descriptor: | 1-{4-amino-3-[6-(cyclopropyloxy)naphthalen-2-yl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}-2-methylpropan-2-ol, Calmodulin-domain protein kinase 1 | | Authors: | Merritt, E.A. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of an Orally Available and Central Nervous System (CNS) Penetrant Toxoplasma gondii Calcium-Dependent Protein Kinase 1 (TgCDPK1) Inhibitor with Minimal Human Ether-a-go-go-Related Gene (hERG) Activity for the Treatment of Toxoplasmosis.
J. Med. Chem., 59, 2016
|
|
1K6Q
 
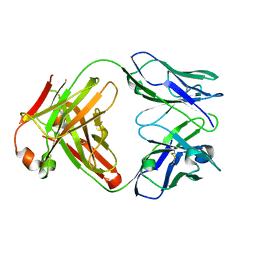 | | Crystal structure of antibody Fab fragment D3 | | Descriptor: | immunoglobulin Fab D3, heavy chain, light chain | | Authors: | Faelber, K, Kelley, R.F, Kirchhofer, D, Muller, Y.A. | | Deposit date: | 2001-10-17 | | Release date: | 2002-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of murine Fab D3 at 2.4 A resolution in comparison with the humanised Fab D3h44 (1.85A) provides structural insight into the humanisation process of the D3 anti-tissue factor antibody
To be Published
|
|
5X6I
 
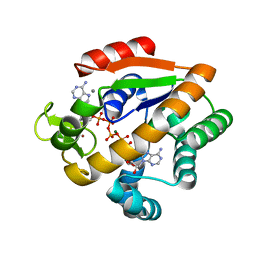 | | Crystal structure of B. subtilis adenylate kinase variant | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, CALCIUM ION, ... | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mutational analyses of psychrophilic and mesophilic adenylate kinases highlight the role of hydrophobic interactions in protein thermal stability.
Struct Dyn., 6, 2019
|
|
5X6J
 
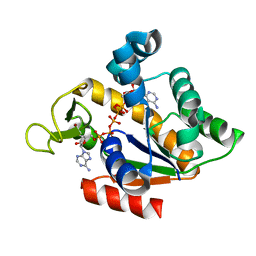 | | Crystal structure of B. globisporus adenylate kinase variant | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Moon, S, Bae, E. | | Deposit date: | 2017-02-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mutational analyses of psychrophilic and mesophilic adenylate kinases highlight the role of hydrophobic interactions in protein thermal stability.
Struct Dyn., 6, 2019
|
|
