1FK3
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH PALMITOLEIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN, PALMITOLEIC ACID | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-09 | | Release date: | 2001-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1FK0
 
 | | STRUCTURAL BASIS OF NON-SPECIFIC LIPID BINDING IN MAIZE LIPID-TRANSFER PROTEIN COMPLEXES WITH CAPRIC ACID REVEALED BY HIGH-RESOLUTION X-RAY CRYSTALLOGRAPHY | | Descriptor: | DECANOIC ACID, FORMIC ACID, NONSPECIFIC LIPID-TRANSFER PROTEIN | | Authors: | Han, G.W, Lee, J.Y, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 2000-08-08 | | Release date: | 2001-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of non-specific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography.
J.Mol.Biol., 308, 2001
|
|
1EOQ
 
 | | ROUS SARCOMA VIRUS CAPSID PROTEIN: C-TERMINAL DOMAIN | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Kingston, R.L, Fitzon-Ostendorp, T, Eisenmesser, E.Z, Schatz, G.W, Vogt, V.M, Post, C.B, Rossmann, M.G. | | Deposit date: | 2000-03-23 | | Release date: | 2000-08-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and self-association of the Rous sarcoma virus capsid protein.
Structure Fold.Des., 8, 2000
|
|
1E91
 
 | | Structure of the complex of the Mad1-Sin3B interaction domains | | Descriptor: | MAD PROTEIN (MAX DIMERIZER), PAIRED AMPHIPATHIC HELIX PROTEIN SIN3B | | Authors: | Spronk, C.A.E.M, Tessari, M, Kaan, A.M, Jansen, J.F.A, Vermeulen, M, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2000-10-04 | | Release date: | 2000-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The MAD1-Sin3B Interaction Involves a Novel Helical Fold
Nat.Struct.Biol., 7, 2000
|
|
9DCN
 
 | |
170D
 
 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA/RNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*T)-R(P*CAR)-D(P*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
171D
 
 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
1B8X
 
 | | GLUTATHIONE S-TRANSFERASE FUSED WITH THE NUCLEAR MATRIX TARGETING SIGNAL OF THE TRANSCRIPTION FACTOR AML-1 | | Descriptor: | PROTEIN (AML-1B) | | Authors: | Tang, L, Guo, B, Van Wijnen, A.J, Lian, J.B, Stein, J.L, Stein, G.S, Zhou, G.W. | | Deposit date: | 1999-02-03 | | Release date: | 1999-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preliminary crystallographic study of glutathione S-transferase fused with the nuclear matrix targeting signal of the transcription factor AML-1/CBF-alpha2.
J.Struct.Biol., 123, 1998
|
|
1ALF
 
 | |
1ALE
 
 | |
3RN3
 
 | | SEGMENTED ANISOTROPIC REFINEMENT OF BOVINE RIBONUCLEASE A BY THE APPLICATION OF THE RIGID-BODY TLS MODEL | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Howlin, B, Moss, D.S, Harris, G.W, Palmer, R.A. | | Deposit date: | 1991-10-30 | | Release date: | 1991-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Segmented anisotropic refinement of bovine ribonuclease A by the application of the rigid-body TLS model.
Acta Crystallogr.,Sect.A, 45, 1989
|
|
3IBK
 
 | |
4HS4
 
 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a Y129N substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-10-29 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
4H6P
 
 | | Crystal structure of a putative chromate reductase from Gluconacetobacter hansenii, Gh-ChrR, containing a R101A substitution. | | Descriptor: | Chromate reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, Y, Robinson, H, Buchko, G.W. | | Deposit date: | 2012-09-19 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.556 Å) | | Cite: | Mechanistic insights of chromate and uranyl reduction by the NADPH-dependent FMN reductase, ChrR, from Gluconacetobacter hansenii
To be Published
|
|
3JZI
 
 | | Crystal structure of biotin carboxylase from E. Coli in complex with benzimidazole series | | Descriptor: | 7-amino-2-[(2-chlorobenzyl)amino]-1-{[(1S,2S)-2-hydroxycycloheptyl]methyl}-1H-benzimidazole-5-carboxamide, Biotin carboxylase | | Authors: | Cheng, C, Shipps, G.W, Yang, Z, Sun, B, Kawahata, N, Soucy, K, Soriano, A, Orth, P, Xiao, L, Mann, P, Black, T. | | Deposit date: | 2009-09-23 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery and optimization of antibacterial AccC inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3KSV
 
 | |
4GXF
 
 | | Role of the biradical intermediate observed during the turnover of SLAC: A two-domain laccase from Streptomyces coelicolor | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Putative copper oxidase, ... | | Authors: | Nederlof, I, Gupta, A, Canters, G.W. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor
J.Am.Chem.Soc., 134, 2012
|
|
4GY4
 
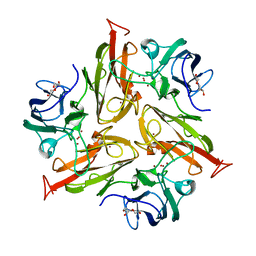 | | Role of the biradical intermediate observed during the turnover of SLAC: A two-domain laccase from Streptomyces coelicolor | | Descriptor: | COPPER (II) ION, OXYGEN ATOM, Putative copper oxidase, ... | | Authors: | Nederlof, I, Gupta, A, Canters, G.W. | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Involvement of Tyr108 in the enzyme mechanism of the small laccase from Streptomyces coelicolor
J.Am.Chem.Soc., 134, 2012
|
|
3LQF
 
 | | Crystal structure of the short-chain dehydrogenase Galactitol-Dehydrogenase (GatDH) of Rhodobacter sphaeroides in complex with NAD and erythritol | | Descriptor: | Galactitol dehydrogenase, MAGNESIUM ION, MESO-ERYTHRITOL, ... | | Authors: | Carius, Y, Christian, H, Faust, A, Kornberger, P, Zander, U, Klink, B.U, Kohring, G.W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into substrate differentiation of the sugar-metabolizing enzyme galactitol dehydrogenase from Rhodobacter sphaeroides D.
J.Biol.Chem., 285, 2010
|
|
4IAQ
 
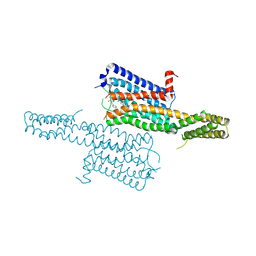 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IB4
 
 | | Crystal structure of the chimeric protein of 5-HT2B-BRIL in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, C, Katritch, V, Han, G.W, Huang, X, Vardy, E, McCorvy, J.D, Jiang, Y, Chu, M, Siu, F.Y, Liu, W, Xu, H.E, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural features for functional selectivity at serotonin receptors.
Science, 340, 2013
|
|
3I4I
 
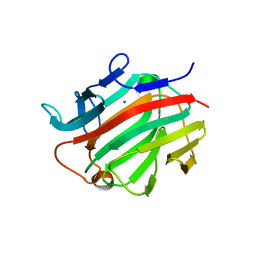 | | Crystal structure of a prokaryotic beta-1,3-1,4-glucanase (lichenase) derived from a mouse hindgut metagenome | | Descriptor: | 1,3-1,4-beta-glucanase, CALCIUM ION | | Authors: | Nakatani, Y, Nalder, T.D, Tannock, G.W, Cutfield, J.F, Jack, R.W, Carne, A. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of a prokaryotic beta-1,3-1,4-glucanase (lichenase) derived from a mouse hindgut metagenome
To be Published
|
|
4H37
 
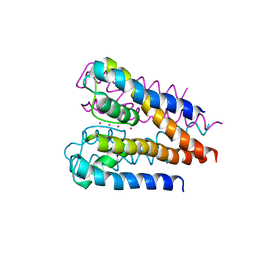 | | Crystal structure of a voltage-gated K+ channel pore domain in a closed state in lipid membranes | | Descriptor: | Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
4H33
 
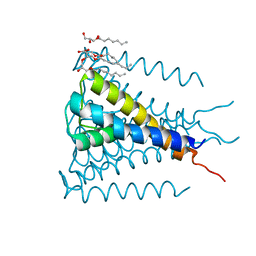 | | Crystal structure of a voltage-gated K+ channel pore module in a closed state in lipid membranes, tetragonal crystal form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
4J5P
 
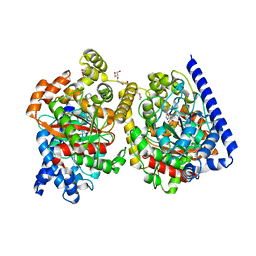 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (1S)-1-{5-[5-(bromomethyl)pyridin-2-yl]-1,3-oxazol-2-yl}-7-phenylheptan-1-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Otrubova, K, Brown, M, McCormick, M.S, Han, G.W, O'Neal, S.T, Cravatt, B.F, Stevens, R.C, Lichtman, A.H, Boger, D.L. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design of Fatty Acid amide hydrolase inhibitors that act by covalently bonding to two active site residues.
J.Am.Chem.Soc., 135, 2013
|
|
