7D28
 
 | |
7D2P
 
 | |
7D2Q
 
 | |
7D2M
 
 | |
1K1R
 
 | | HETERODUPLEX OF CHIRALLY PURE R-METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(RMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenkova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
4ARE
 
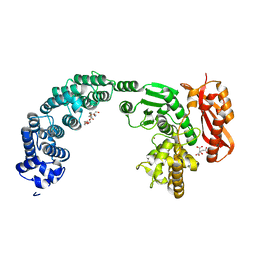 | | Crystal structure of the collagenase Unit of collagenase G from Clostridium histolyticum at 2.19 angstrom resolution. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, COLLAGENASE G, ... | | Authors: | Eckhard, U, Brandstetter, H. | | Deposit date: | 2012-04-23 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for Activity Regulation and Substrate Preference of Clostridial Collagenases G, H, and T.
J.Biol.Chem., 288, 2013
|
|
7QDT
 
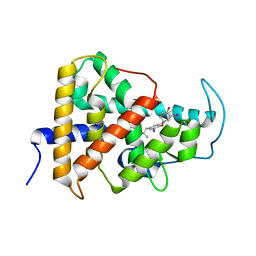 | | Crystal structure of a mutant (P393GX) Thyroid Receptor Alpha ligand binding domain designed to model dominant negative human mutations. | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Isoform Alpha-1 of Thyroid hormone receptor alpha | | Authors: | Romartinez-Alonso, B, Fairall, L, Agostini, M, Chatterjee, K, Schwabe, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Guided Approach to Relieving Transcriptional Repression in Resistance to Thyroid Hormone alpha.
Mol.Cell.Biol., 42, 2022
|
|
6I5R
 
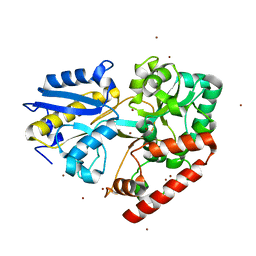 | | BlMnBP1 binding protein of an ABC transporter from Bifidobacterium animalis subsp. lactis ATCC27673 in complex with mannobiose | | Descriptor: | Sugar ABC transporter substrate-binding protein, BlMnBP1, ZINC ION, ... | | Authors: | Abou Hachem, M, Ejby, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-11-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
5MGT
 
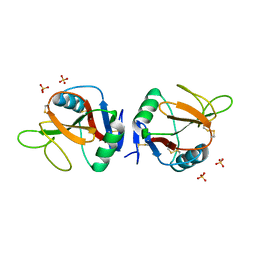 | | Complex of human NKR-P1 and LLT1 in deglycosylated forms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, CHLORIDE ION, ... | | Authors: | Blaha, J, Skalova, T, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
5MGR
 
 | | Human receptor NKR-P1 in glycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
6I5V
 
 | | BlMnBP1 binding protein of an ABC transporter from Bifidobacterium animalis subsp. lactis ATCC27673 in complex with mannotriose | | Descriptor: | Solute Binding Protein, BlMnBP1 in complex with mannotrisoe, ZINC ION, ... | | Authors: | Abou Hachem, M, Ejby, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-11-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
2Y6I
 
 | |
6I5W
 
 | | BlMnBP1 binding protein of an ABC transporter from Bifidobacterium animalis subsp. lactis ATCC27673 in complex with mannobiose | | Descriptor: | ACETATE ION, Solute Binding Protein BlMnBP1, Blac_00780 in complex with mannopentaose, ... | | Authors: | Abou Hachem, M, Ejby, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-11-14 | | Release date: | 2019-04-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
5MGS
 
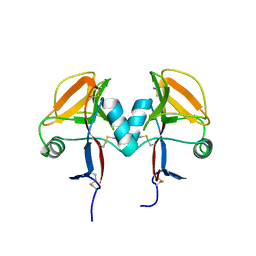 | | Human receptor NKR-P1 in deglycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1 | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
3FLI
 
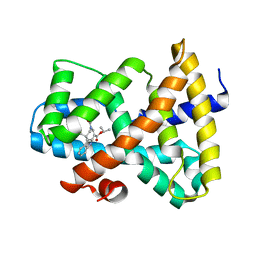 | | Discovery of XL335, a Highly Potent, Selective and Orally-Active Agonist of the Farnesoid X Receptor (FXR) | | Descriptor: | 1-methylethyl 3-[(3,4-difluorophenyl)carbonyl]-1,1-dimethyl-1,2,3,6-tetrahydroazepino[4,5-b]indole-5-carboxylate, Bile acid receptor | | Authors: | Foster, P.G, Stout, T.J. | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of XL335 (WAY-362450), a highly potent, selective, and orally active agonist of the farnesoid X receptor (FXR).
J.Med.Chem., 52, 2009
|
|
6FUV
 
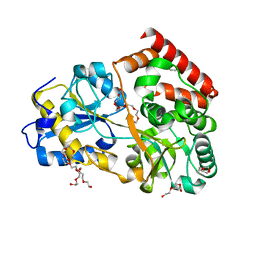 | | Structure of a manno-oligosaccharide specific solute binding protein, BlMnBP2 from Bifidobacterium animalis subsp. lactis ATCC 27673 in complex with mannotriose | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETATE ION, Solute Binding Protein, ... | | Authors: | Ejby, M, Abou Hachem, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-02-27 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Two binding proteins of the ABC transporter that confers growth of Bifidobacterium animalis subsp. lactis ATCC27673 on beta-mannan possess distinct manno-oligosaccharide-binding profiles.
Mol.Microbiol., 112, 2019
|
|
1H80
 
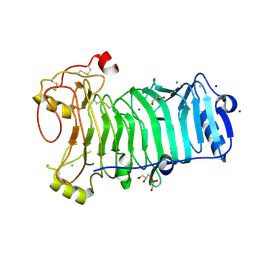 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE- 3,6-ANHYDRO-D-GALACTOSE-2-SULFATE 4 GALACTOHYDROLASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2001-01-22 | | Release date: | 2001-11-27 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Iota-Carrageenase of Alteromonas Fortis. A Beta-Helix Fold-Containing Enzyme for the Degradation of a Highly Polyanionic Polysaccharide
J.Biol.Chem., 276, 2001
|
|
5O85
 
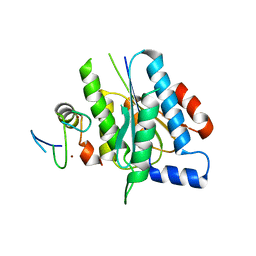 | | p34-p44 complex | | Descriptor: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, ZINC ION | | Authors: | Radu, L, Poterszman, A. | | Deposit date: | 2017-06-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
1EAJ
 
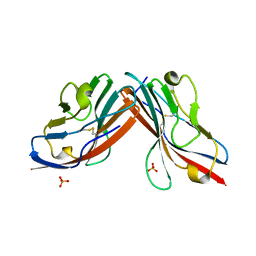 | |
7MHC
 
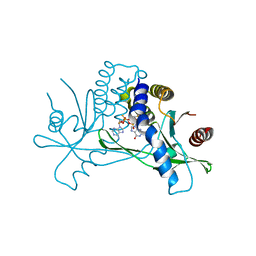 | | Structure of human STING in complex with MK-1454 | | Descriptor: | (2R,5R,7R,8S,10R,12aR,14R,15S,15aR,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-14-(6-amino-9H-purin-9-yl)-15,16-difluoro-2,10-bis(sulfanyl)octahydro-2H,10H,12H-5,8-methano-2lambda~5~,10lambda~5~-furo[3,2-l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A kinase-cGAS cascade to synthesize a therapeutic STING activator.
Nature, 603, 2022
|
|
6EXM
 
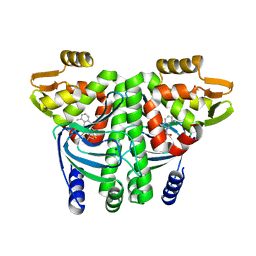 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK202) | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Grundstrom, C, Begum, A, Hall, M, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUT
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK67) | | Descriptor: | (3~{R})-8-cyclopropyl-7-[(4-methylnaphthalen-1-yl)methyl]-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUZ
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK37) | | Descriptor: | (3~{R})-8-methoxy-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EXL
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - folded HTH motif | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUU
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK29) | | Descriptor: | (3~{R})-8-cyclopropyl-5-oxidanylidene-7-(quinolin-3-ylmethyl)-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
