9KMG
 
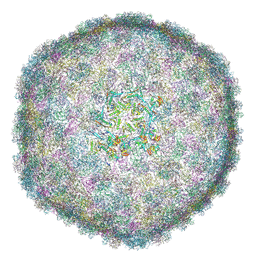 | | Cryo-EM Structure of Bacteriophage FCWL1 Capsid | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Cai, C, Wang, Y, Liu, Y, Shao, Q, Wang, A, Fang, Q. | | Deposit date: | 2024-11-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of a T1-like siphophage reveal capsid stabilization mechanisms and high structural similarities with a myophage.
Structure, 33, 2025
|
|
1D4N
 
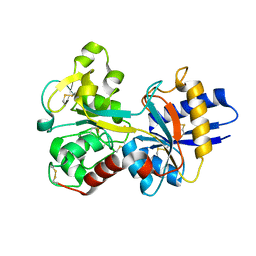 | | HUMAN SERUM TRANSFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, TRANSFERRIN | | Authors: | Yang, H.-W, MacGillivray, R.T.A, Chen, J, Luo, Y, Wang, Y, Brayer, G.D, Mason, A, Woodworth, R.C, Murphy, M.E.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two mutants (K206Q, H207E) of the N-lobe of human transferrin with increased affinity for iron.
Protein Sci., 9, 2000
|
|
1D3K
 
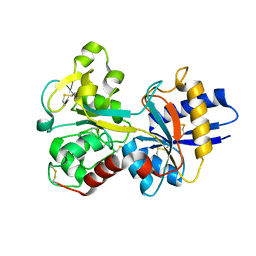 | | HUMAN SERUM TRANSFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Yang, H.-W, MacGillivray, R.T.A, Chen, J, Luo, Y, Wang, Y, Brayer, G.D, Mason, A, Woodworth, R.C, Murphy, M.E.P. | | Deposit date: | 1999-09-29 | | Release date: | 2000-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two mutants (K206Q, H207E) of the N-lobe of human transferrin with increased affinity for iron.
Protein Sci., 9, 2000
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
4KPY
 
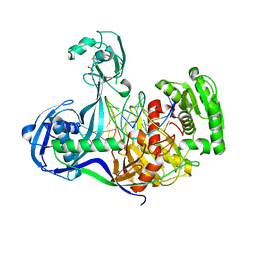 | | DNA binding protein and DNA complex structure | | Descriptor: | DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*CP*C)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3'), DNA (5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3'), ... | | Authors: | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9KPN
 
 | | Crystal structure of KRAS-G12C in complex with Compound 20 (JAB-20) | | Descriptor: | 7-[2-azanyl-3,4,5,6-tetrakis(fluoranyl)phenyl]-6-chloranyl-1-(4-methyl-2-propan-2-yl-pyridin-3-yl)-2-oxidanylidene-4-(4-prop-2-enoylpiperazin-1-yl)-1,8-naphthyridine-3-carbonitrile, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, S, Dang, C, Fan, X, Zhang, W, Wang, P, Qian, D, Wang, Y, Li, A. | | Deposit date: | 2024-11-23 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Design, Structure Optimization, and Preclinical Characterization of JAB-21822, a Covalent Inhibitor of KRAS G12C.
J.Med.Chem., 68, 2025
|
|
9KPM
 
 | | Crystal structure of KRAS-G12C in complex with compound 16 (JAB-16) | | Descriptor: | 7-[2-azanyl-3,5-bis(chloranyl)-6-fluoranyl-phenyl]-6-chloranyl-1-(4-methyl-2-propan-2-yl-pyridin-3-yl)-2-oxidanylidene-4-(4-prop-2-enoylpiperazin-1-yl)-1,8-naphthyridine-3-carbonitrile, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Li, S, Dang, C, Fan, X, Zhang, W, Wang, P, Qian, D, Wang, Y, Li, A. | | Deposit date: | 2024-11-23 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design, Structure Optimization, and Preclinical Characterization of JAB-21822, a Covalent Inhibitor of KRAS G12C.
J.Med.Chem., 68, 2025
|
|
5XAF
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
5GNF
 
 | | Crystal structure of anti-CRISPR protein AcrF3 | | Descriptor: | CALCIUM ION, Uncharacterized protein AcrF3 | | Authors: | Wang, J, Wang, Y. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
8DOF
 
 | | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044 | | Descriptor: | (2R)-2-cyclohexyl-2-[(4-{[5-(propan-2-yl)-1H-pyrazol-3-yl]amino}-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]ethan-1-ol, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Horanyi, P.S, Wang, Y, Todd, M.H, Abendroth, J, Edwards, T, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-07-13 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pseudomonas aeruginosa MurC with WYH9-2-P - OSA_001044
To Be Published
|
|
1MRH
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1YT3
 
 | | Crystal Structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing | | Descriptor: | Ribonuclease D, SULFATE ION, ZINC ION | | Authors: | Zuo, Y, Wang, Y, Malhotra, A. | | Deposit date: | 2005-02-09 | | Release date: | 2005-08-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Escherichia coli RNase D, an Exoribonuclease Involved in Structured RNA Processing
Structure, 13, 2005
|
|
4H75
 
 | | Crystal structure of human Spindlin1 in complex with a histone H3K4(me3) peptide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Histone H3, ... | | Authors: | Yang, N, Wang, W, Wang, Y, Wang, M, Zhao, Q, Rao, Z, Zhu, B, Xu, R.M. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
1MRI
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
1MRJ
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1PDQ
 
 | | Polycomb chromodomain complexed with the histone H3 tail containing trimethyllysine 27. | | Descriptor: | Histone H3.3, Polycomb protein | | Authors: | Fischle, W, Wang, Y, Jacobs, S.A, Kim, Y, Allis, C.D, Khorasanizadeh, S. | | Deposit date: | 2003-05-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains
Genes Dev., 17, 2003
|
|
1MS6
 
 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | Descriptor: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | Authors: | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-04-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
8HYR
 
 | |
5FBJ
 
 | | Complex structure of JMJD5 and substrate | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, ... | | Authors: | Liu, H.L, Wang, Y, Wang, C, Zhang, G.Y. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | to be published
To Be Published
|
|
3GO6
 
 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose and AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOKINASE RBSK, ... | | Authors: | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | Deposit date: | 2009-03-18 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
