8BSM
 
 | | Human GLS in complex with compound 18 | | Descriptor: | 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial 65 kDa chain, ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.782 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSK
 
 | | Human GLS in complex with compound 3 | | Descriptor: | Glutaminase kidney isoform, mitochondrial 65 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide), ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSL
 
 | | Human GLS in complex with compound 12 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, ~{N}-[5-[[(3~{R})-1-(5-azanyl-1,3,4-thiadiazol-2-yl)pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]-2-phenyl-ethanamide | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSN
 
 | | Human GLS in complex with compound 27 | | Descriptor: | (2~{S})-2-methoxy-2-phenyl-~{N}-[5-[[(3~{R})-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial 65 kDa chain, ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8HPU
 
 | |
8HQ7
 
 | |
8HPF
 
 | |
8HPV
 
 | |
8HP9
 
 | |
8HPQ
 
 | |
4LA3
 
 | |
7YR5
 
 | |
4G9G
 
 | |
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
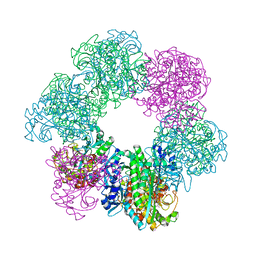 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
4HSU
 
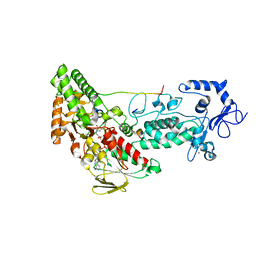 | | Crystal structure of LSD2-NPAC with H3(1-26)in space group P21 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3, Lysine-specific histone demethylase 1B, ... | | Authors: | Chen, F, Dong, Z, Fang, J, Xu, Y. | | Deposit date: | 2012-10-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural insight into substrate recognition by histone demethylase LSD2/KDM1b.
Cell Res., 23, 2013
|
|
7CKM
 
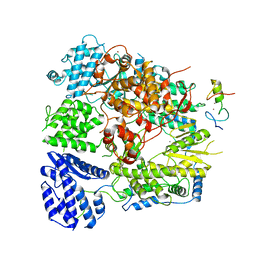 | | Structure of Machupo virus polymerase bound to Z matrix protein (monomeric complex) | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
7CKL
 
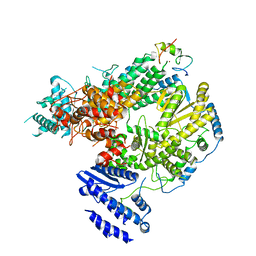 | | Structure of Lassa virus polymerase bound to Z matrix protein | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
7CM9
 
 | | DMSP lyase DddX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DMSP lyase, SULFATE ION | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-07-25 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA.
Elife, 10, 2021
|
|
8VTT
 
 | | Meis1 homeobox domain bound to neomycin fragment | | Descriptor: | Homeobox protein Meis1, RIBOSTAMYCIN, SULFATE ION | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N, Sadek, H.A. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
8VTS
 
 | | Meis1 homeobox domain bound to paromomycin fragment | | Descriptor: | 1,2-ETHANEDIOL, Homeobox protein Meis1, ISOPROPYL ALCOHOL, ... | | Authors: | Tomchick, D.R, Ahmed, M.S, Nguyen, N.U.N. | | Deposit date: | 2024-01-27 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of FDA-approved drugs that induce heart regeneration in mammals.
Nat Cardiovasc Res, 3, 2024
|
|
7ECW
 
 | | The Csy-AcrIF14-dsDNA complex | | Descriptor: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ECV
 
 | | The Csy-AcrIF14 complex | | Descriptor: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7DU0
 
 | | Structure of an type I-F anti-crispr protein | | Descriptor: | AcrIF14 | | Authors: | Teng, G, Yue, F. | | Deposit date: | 2021-01-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
