6CZ1
 
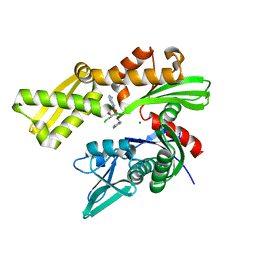 | | Crystal structure of ATPase domain of Human GRP78 bound to Ver155008 | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Endoplasmic reticulum chaperone BiP, MAGNESIUM ION | | Authors: | Antoshchenko, T, Chen, Y, Hughes, S, Park, H. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic selection of adenosine analogs that fit the mold of the active site of human GRP78 and beyond
To be Published
|
|
7RD0
 
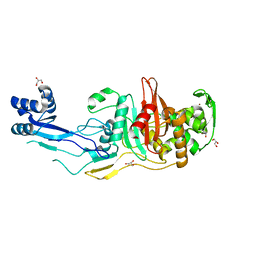 | |
7RCW
 
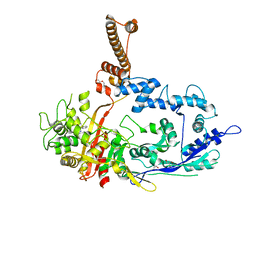 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCZ
 
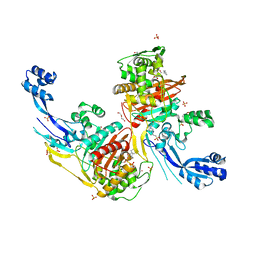 | | Crystal structure of C. difficile SpoVD in complex with ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCY
 
 | | Crystal structure of C. difficile penicillin-binding protein 2 in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein, ZINC ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2021-07-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique class of Zn 2+ -binding serine-based PBPs underlies cephalosporin resistance and sporogenesis in Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7RCX
 
 | |
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
5XP5
 
 | | C-Src in complex with ATP-Chf | | Descriptor: | MAGNESIUM ION, Proto-oncogene tyrosine-protein kinase Src, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[(S)-fluoranyl-[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Duan, Y, Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Remarkably Stereospecific Utilization of ATP alpha , beta-Halomethylene Analogues by Protein Kinases.
J. Am. Chem. Soc., 139, 2017
|
|
1C76
 
 | | STAPHYLOKINASE (SAK) MONOMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Staphylokinase Dimer Offers New Clue to Reduction of Immunogenicity
To be published
|
|
4ZHT
 
 | | Crystal structure of UDP-GlcNAc 2-epimerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-mannopyranose, Bifunctional UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase, CYTIDINE-5'-MONOPHOSPHATE-5-N-ACETYLNEURAMINIC ACID, ... | | Authors: | Chen, S.C, Yang, C.S, Ko, T.P, Chen, Y. | | Deposit date: | 2015-04-27 | | Release date: | 2016-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Mechanism and inhibition of human UDP-GlcNAc 2-epimerase, the key enzyme in sialic acid biosynthesis.
Sci Rep, 6, 2016
|
|
9J4I
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) in compex with fruetosyl nystose (GF4) | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-[alpha-D-glucopyranose-(1-2)]beta-D-fructofuranose | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4K
 
 | | Crystal structure of GH9l Inulinfructotransferases (IFTase) in complex with GF2 | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4J
 
 | | Crystal structure of GH9l Inulin fructotransferases(IFTase)incomplex with nystose(F3) | | Descriptor: | DFA-III-forming inulin fructotransferase, beta-D-fructofuranose, beta-D-fructofuranose-(1-1)-beta-D-fructofuranose, ... | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
9J4L
 
 | | Crystal structure of GH9l Inulin fructotransferases (IFTase) | | Descriptor: | DFA-III-forming inulin fructotransferase | | Authors: | Chen, G, Wang, Z.X, Yang, Y.Q, Li, Y.G, Zhang, T, Ouyang, S.Y, Zhang, L, Chen, Y, Ruan, X.L, Miao, M. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Elucidation of the mechanism underlying the sequential catalysis of inulin by fructotransferase.
Int.J.Biol.Macromol., 277, 2024
|
|
1ZKL
 
 | | Multiple Determinants for Inhibitor Selectivity of Cyclic Nucleotide Phosphodiesterases | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High-affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Wang, H, Liu, Y, Chen, Y, Robinson, H, Ke, H. | | Deposit date: | 2005-05-03 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7
J.Biol.Chem., 280, 2005
|
|
1VBT
 
 | | Structure of cyclophilin complexed with sulfur-substituted tetrapeptide AAPF | | Descriptor: | CYCLOPHILIN A, SULFUR-SUBSTITUTED TETRAPEPTIDE | | Authors: | Zhao, Y, Chen, Y, Schutkowski, M, Fischer, G, Ke, H. | | Deposit date: | 1998-06-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into Conversion of Substrate to Inhibitor
To be Published
|
|
1XKL
 
 | | Crystal Structure of Salicylic Acid-binding Protein 2 (SABP2) from Nicotiana tabacum, NESG Target AR2241 | | Descriptor: | 2-AMINO-4H-1,3-BENZOXATHIIN-4-OL, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Chen, Y, Chiang, Y, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4KYK
 
 | | Crystal structure of mouse glyoxalase I complexed with indomethacin | | Descriptor: | INDOMETHACIN, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
2B34
 
 | | Structure of MAR1 Ribonuclease from Caenorhabditis elegans | | Descriptor: | MAR1 Ribonuclease | | Authors: | Schormann, N, Karpova, E, Li, S, Symersky, J, Zhang, Y, Lu, S, Zhou, Q, Lin, G, Cao, Z, Luo, M, Qiu, S, Luan, C.-H, Luo, D, Huang, W, Shang, Q, McKinstry, A, An, J, Tsao, J, Carson, M, Stinnett, M, Chen, Y, Johnson, D, Gary, R, Arabshahi, A, Bunzel, R, Bray, T, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-09-19 | | Release date: | 2005-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structure of MAR1 Ribonuclease from Caenorhabditis elegans
To be Published
|
|
4KYH
 
 | | Crystal structure of mouse glyoxalase I complexed with zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Lactoylglutathione lyase, ZINC ION | | Authors: | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
6L0Z
 
 | | The crystal structure of Salmonella enterica sugar-binding protein MalE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-anhydro-D-glucitol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Salmonella enterica sugar-binding protein MalE
To Be Published
|
|
6L19
 
 | | The crystal structure of competence or damage-inducible protein from Enterobacter asburiae | | Descriptor: | CHLORIDE ION, GLYCEROL, PncC family amidohydrolase, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The crystal structure of Competence or damage-inducible protein from Enterobacter asburiae
To Be Published
|
|
6L1K
 
 | | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum | | Descriptor: | NADH-dependent butanol dehydrogenase A, PHOSPHATE ION | | Authors: | Lan, J, Shang, F, Liu, W, Xu, Y, Chen, Y. | | Deposit date: | 2019-09-29 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of NADH-dependent butanol dehydrogenase A from Fusobacterium nucleatum
To Be Published
|
|
3E23
 
 | | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299 | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, uncharacterized protein RPA2492 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-05 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299
To be Published
|
|
3E0H
 
 | | Crystal structure of an uncharacterized protein from Chlorobium tepidum. NorthEast Structural Genomics target CtR107. | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of an uncharacterized protein from Chlorobium tepidum. NorthEast Structural Genomics target CtR107.
To be Published
|
|
