6X5H
 
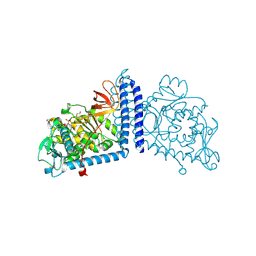 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP | | Descriptor: | 1,2-ETHANEDIOL, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
6WMM
 
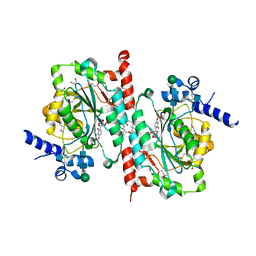 | |
6X5U
 
 | | Human Alpha-1,6-fucosyltransferase (FUT8) bound to GDP and NM5M2-Asn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterizing human alpha-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases.
J.Biol.Chem., 295, 2020
|
|
5FBH
 
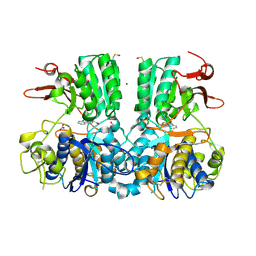 | | Crystal structure of the extracellular domain of human calcium sensing receptor with bound Gd3+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
5FBK
 
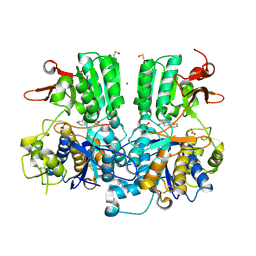 | | Crystal structure of the extracellular domain of human calcium sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
6JYJ
 
 | | Crystal structure of FAM46B (TENT5B) | | Descriptor: | CITRATE ANION, Terminal nucleotidyltransferase 5B | | Authors: | Zhang, H, Hu, J.L, Gao, S. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.693199 Å) | | Cite: | FAM46B is a prokaryotic-like cytoplasmic poly(A) polymerase essential in human embryonic stem cells.
Nucleic Acids Res., 48, 2020
|
|
5VCM
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound UDP and Manganese | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VCR
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with bound uranium dioxide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, CHLORIDE ION, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5VCS
 
 | | Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase with Bound Acceptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase, ... | | Authors: | Sanders, J.H, Kadirvelraj, R, Wood, Z.A. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | HumanN-acetylglucosaminyltransferase II substrate recognition uses a modular architecture that includes a convergent exosite.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6K94
 
 | |
6K93
 
 | |
5GOE
 
 | | Truncated mitofusin-1, GDP-bound | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitofusin-1 | | Authors: | Cao, Y.L, Gao, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion
Nature, 542, 2017
|
|
5GO4
 
 | | Truncated mitofusin-1, nucleotide-free | | Descriptor: | Mitofusin-1 | | Authors: | Cao, Y.L, Gao, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion
Nature, 542, 2017
|
|
5GOF
 
 | | Truncated mitofusin-1, GTP-bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitofusin-1, ... | | Authors: | Cao, Y.L, Gao, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion
Nature, 542, 2017
|
|
5GOM
 
 | | Truncated mitofusin-1, transition-like state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GUANOSINE-5'-DIPHOSPHATE, Mitofusin-1 | | Authors: | Cao, Y.L, Gao, S. | | Deposit date: | 2016-07-27 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion
Nature, 542, 2017
|
|
6APJ
 
 | | Crystal Structure of human ST6GALNAC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 | | Authors: | Forouhar, F, Moremen, K.W, Northeast Structural Genomics Consortium (NESG), Tong, L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Expression system for structural and functional studies of human glycosylation enzymes.
Nat. Chem. Biol., 14, 2018
|
|
6APL
 
 | | Crystal Structure of human ST6GALNAC2 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Forouhar, F, Moremen, K.W, Northeast Structural Genomics Consortium (NESG), Tong, L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Expression system for structural and functional studies of human glycosylation enzymes.
Nat. Chem. Biol., 14, 2018
|
|
6CCI
 
 | | The Crystal Structure of XOAT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2018-02-07 | | Release date: | 2019-02-20 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanism of Polysaccharide Acetylation by the Arabidopsis XylanO-acetyltransferase XOAT1.
Plant Cell, 32, 2020
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5YDG
 
 | | Crystal structure of the Arabidopsis thaliana chloroplast RNA editing factors 2(MORF2) | | Descriptor: | Multiple organellar RNA editing factor 2, chloroplastic | | Authors: | Wang, X, Yang, J.Y, Wang, Y.L, Gao, Y.S. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of the chloroplast RNA editing factor MORF2
Biochem. Biophys. Res. Commun., 495, 2018
|
|
