8GC7
 
 | |
2XXN
 
 | | Structure of the vIRF4-HAUSP TRAF domain complex | | Descriptor: | K10, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 7 | | Authors: | Choi, W.C, Hwang, J, Kim, M.H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bilateral Inhibition of Hausp Deubiquitinase by a Viral Interferon Regulatory Factor Protein
Nat.Struct.Mol.Biol., 18, 2011
|
|
7C13
 
 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin, Peptide methionine sulfoxide reductase MsrA | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7C12
 
 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7C10
 
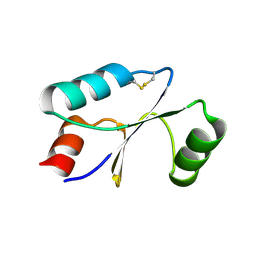 | | Dithiol cGrx1 | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
8GSA
 
 | |
4GFX
 
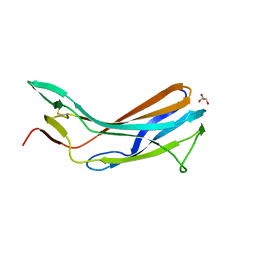 | | Crystal structure of the N-terminal domain of TXNIP | | Descriptor: | GLYCEROL, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2012-08-04 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein.
Nat Commun, 5, 2014
|
|
5YCX
 
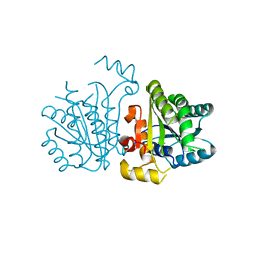 | |
4LL4
 
 | | The structure of the TRX and TXNIP complex | | Descriptor: | Thioredoxin, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein
Nat Commun, 5, 2014
|
|
4LL1
 
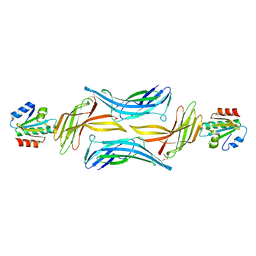 | | The structure of the TRX and TXNIP complex | | Descriptor: | Thioredoxin, Thioredoxin-interacting protein | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the negative regulation of thioredoxin by thioredoxin-interacting protein
Nat Commun, 5, 2014
|
|
5YCS
 
 | |
5YCR
 
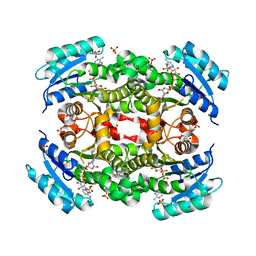 | |
5YCV
 
 | |
7WBN
 
 | | PDB structure of RevCC | | Descriptor: | RevCC | | Authors: | Han, S, Kim, D, Kaur, M, Lim, Y.B, Barnwal, R.P. | | Deposit date: | 2021-12-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pseudo-Isolated alpha-Helix Platform for the Recognition of Deep and Narrow Targets.
J.Am.Chem.Soc., 144, 2022
|
|
6M2D
 
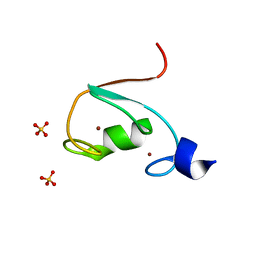 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
6M2C
 
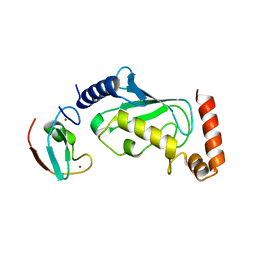 | |
7BOL
 
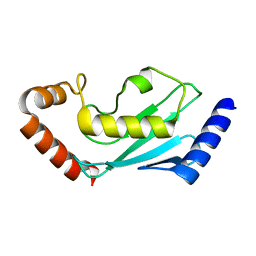 | | ubiquitin-conjugating enzyme, Ube2D2 | | Descriptor: | Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
7D16
 
 | | NERP-2 in a DPC solution | | Descriptor: | Neurosecretory protein VGF | | Authors: | Park, O, Cheong, C, Jeon, Y. | | Deposit date: | 2020-09-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of neuroendocrine regulatory peptide-2 in membrane-mimicking environments.
Peptide Science, 2020
|
|
2KSE
 
 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor QseC, Center for Structures of Membrane Proteins (CSMP) target 4311C | | Descriptor: | Sensor protein qseC | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7D13
 
 | | NERP-2 in a HFIP solution | | Descriptor: | Neurosecretory protein VGF | | Authors: | Park, O, Cheong, C, Jeon, Y. | | Deposit date: | 2020-09-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of neuroendocrine regulatory peptide-2 in membrane-mimicking environments.
Peptide Science, 2020
|
|
7F27
 
 | | Crystal structure of polyketide ketosynthase | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
7F28
 
 | | Crystal structure of a bacterial ketosynthase | | Descriptor: | Ketoacyl_synth_N domain-containing protein, Putative 3-oxoacyl-[ACP] synthase FabV | | Authors: | Lee, W.C, Kim, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
2KSF
 
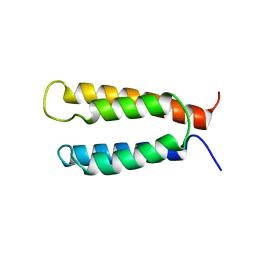 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor KdpD, Center for Structures of Membrane Proteins (CSMP) target 4312C | | Descriptor: | Sensor protein kdpD | | Authors: | Maslennikov, I, Klammt, C, Kefala, G, Okamura, M, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-01-03 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5Z9G
 
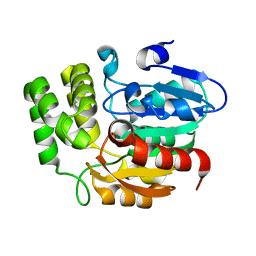 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5Z9H
 
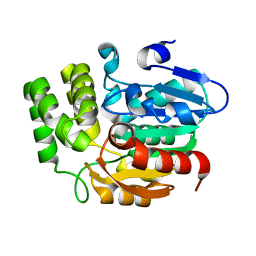 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
