3UA3
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 in complex with SAH | | Descriptor: | Protein arginine N-methyltransferase 5, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | Deposit date: | 2011-10-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3UA4
 
 | | Crystal Structure of Protein Arginine Methyltransferase PRMT5 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 5 | | Authors: | Sun, L, Wang, M, Lv, Z, Yang, N, Liu, Y, Bao, S, Gong, W, Xu, R.M. | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural insights into protein arginine symmetric dimethylation by PRMT5
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4JPN
 
 | | Bacteriophage phiX174 H protein residues 143-221 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
4JPP
 
 | | Bacteriophage phiX174 H protein residues 143-282 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
8JWE
 
 | |
3SSA
 
 | | Crystal structure of subunit B mutant N157T of the A1AO ATP synthase | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sundararaman, L, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-07-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of subunit A mutants H156A, N157A and N157T of the A1AO ATP synthase
To be Published
|
|
7RWE
 
 | | Crystal structure of CDK2 liganded with compound GPHR787 | | Descriptor: | 5-nitro-2-[(3-phenylpropyl)amino]benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7RXO
 
 | | Crystal structure of CDK2 liganded with compound WN333 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(methoxycarbonyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S4T
 
 | | Crystal structure of CDK2 liganded with compound EF2252 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(6-chloro-1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S7A
 
 | | Crystal structure of CDK2 liganded with compound EF3019 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(2H-indazol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S85
 
 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
4XOH
 
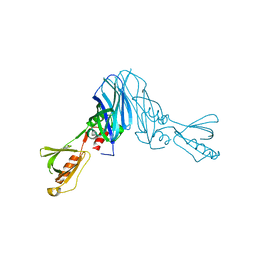 | | Mechanistic insights into anchorage of the contractile ring from yeast to humans | | Descriptor: | Division mal foutue 1 protein | | Authors: | Chen, Z, Wu, J.-Q, Wang, J, Guan, R, Sun, L, Lee, I.-J, Liu, Y, Chen, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Mechanistic insights into the anchorage of the contractile ring by anillin and mid1
Dev.Cell, 33, 2015
|
|
6IJZ
 
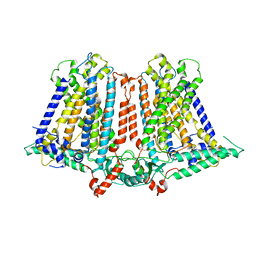 | | Structure of a plant cation channel | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Sun, L, Wang, J, Liu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2.
Nat Commun, 9, 2018
|
|
7RWF
 
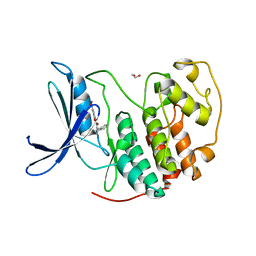 | | Crystal structure of CDK2 in complex with TW8672 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7S84
 
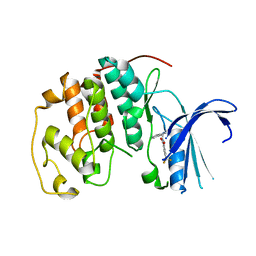 | | Crystal structure of CDK2 liganded with compound TW8972 | | Descriptor: | 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7MPH
 
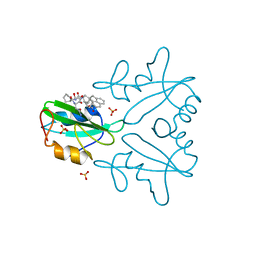 | | GRB2 SH2 Domain with Compound 7 | | Descriptor: | (4-{(10R,11E,14S,18S)-18-(2-amino-2-oxoethyl)-14-[(naphthalen-1-yl)methyl]-8,17,20-trioxo-7,16,19-triazaspiro[5.14]icos-11-en-10-yl}phenyl)acetic acid, 1,2-ETHANEDIOL, Growth factor receptor-bound protein 2, ... | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structural characterization of a monocarboxylic inhibitor for GRB2 SH2 domain.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
5U6K
 
 | |
7Y9V
 
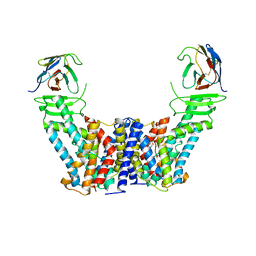 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the IAA-bound state | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9U
 
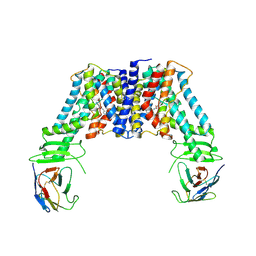 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the NPA-bound state | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7Y9T
 
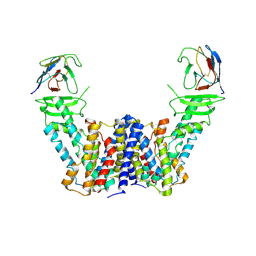 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the apo state | | Descriptor: | Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2022-06-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into auxin recognition and efflux by Arabidopsis PIN1.
Nature, 609, 2022
|
|
7YKS
 
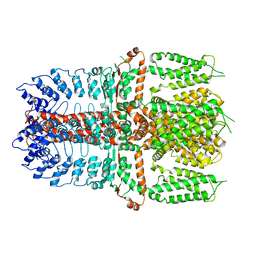 | |
7YKR
 
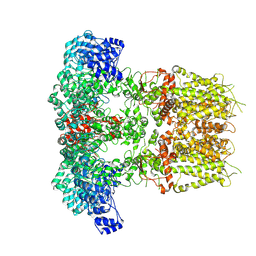 | |
8J2G
 
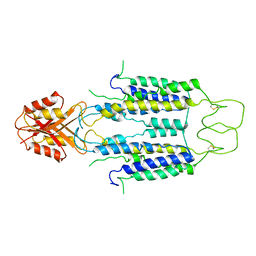 | | Human ZnT1-D47N/D255N mutant | | Descriptor: | Proton-coupled zinc antiporter SLC30A1 | | Authors: | Sun, L, Liu, X, Sun, C, He, B. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the calcium-coupled zinc export of human ZnT1.
Sci Adv, 10, 2024
|
|
8JH5
 
 | | Structure of the auxin exporter PIN1 in Arabidopsis thaliana in the Naproxen-bound state | | Descriptor: | (2S)-2-(6-methoxynaphthalen-2-yl)propanoic acid, Auxin efflux carrier component 1, nanobody | | Authors: | Sun, L, Liu, X, Yang, Z, Xia, J. | | Deposit date: | 2023-05-22 | | Release date: | 2024-05-29 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chemical inhibition of Arabidopsis PIN-FORMED auxin transporters by the anti-inflammatory drug naproxen.
Plant Commun., 4, 2023
|
|
8GQ6
 
 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | Descriptor: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | Authors: | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
