6I5H
 
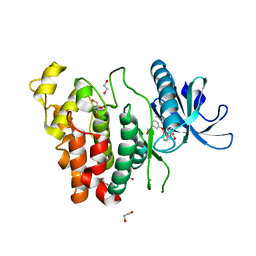 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5L3L
 
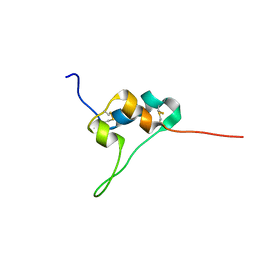 | | D11 bound IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L3M
 
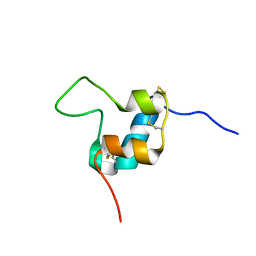 | | D11 bound [S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
5L3N
 
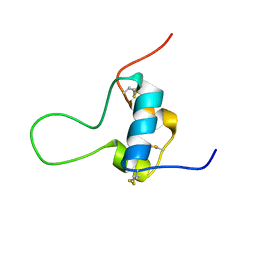 | | D11 bound [N29, S39_PQ]-IGF-II | | Descriptor: | Insulin-like growth factor II | | Authors: | Hexnerova, R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Probing Receptor Specificity by Sampling the Conformational Space of the Insulin-like Growth Factor II C-domain.
J.Biol.Chem., 291, 2016
|
|
6I5L
 
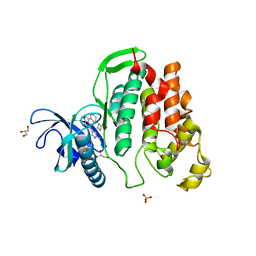 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN316 (derivative of compound 12h) | | Descriptor: | 3-(3-cyclobutylphenyl)-5-(1-methylpyrazol-4-yl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6I5I
 
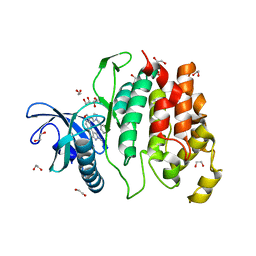 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound 12h | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-[1-(phenylmethyl)pyrazol-4-yl]furo[3,2-b]pyridine, Dual specificity protein kinase CLK1 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6I5K
 
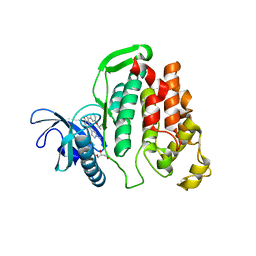 | | Crystal structure of CLK1 in complexed with furo[3,2-b]pyridine compound VN345 (derivative of compound 12h) | | Descriptor: | 5-(1-methylpyrazol-4-yl)-3-(3-propan-2-yloxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, GLYCEROL, ... | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
2N2X
 
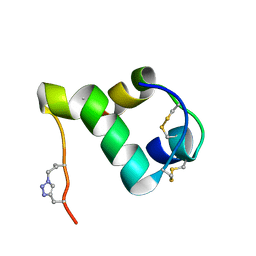 | |
2N2W
 
 | |
2MPG
 
 | | Solution structure of the [AibB8,LysB28,ProB29]-insulin analogue | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Kosinova, L, Jiracek, J, Zakova, L, Veverka, V. | | Deposit date: | 2014-05-17 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Insight into the structural and biological relevance of the T/R transition of the N-terminus of the B-chain in human insulin.
Biochemistry, 53, 2014
|
|
2N2V
 
 | |
5BOQ
 
 | | Human insulin with intra-chain chemical crosslink between modified B24 and B29 | | Descriptor: | Insulin, SULFATE ION | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2015-05-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational steering of insulin binding specificity by intra-chain chemical crosslinking.
Sci Rep, 6, 2016
|
|
5BPO
 
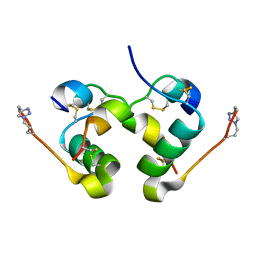 | |
5BQQ
 
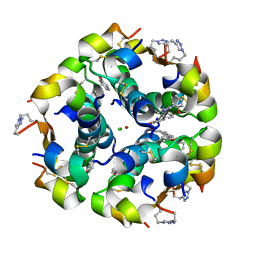 | | Human insulin with intra-chain chemical crosslink between modified B27 and B30 | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2015-05-29 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Rational steering of insulin binding specificity by intra-chain chemical crosslinking.
Sci Rep, 6, 2016
|
|
8OW7
 
 | | Crystal structure of Tannerella forsythia sugar kinase K1058 in complex with N-acetylmuramic acid (MurNAc) | | Descriptor: | N-acetyl-beta-muramic acid, N-acetylglucosamine kinase, SULFATE ION | | Authors: | Stasiak, A.C, Gogler, K, Fink, P, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OW9
 
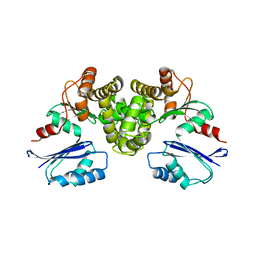 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK in complex with N-acetylmuramic acid (MurNAc) | | Descriptor: | N-acetyl-beta-muramic acid, Putative novel MurNAc kinase | | Authors: | Stasiak, A.C, Gogler, K, Fink, P, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OQW
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK | | Descriptor: | ATPase, GLYCEROL, SULFATE ION | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OQX
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK with a phosphate analogue | | Descriptor: | 1,2-ETHANEDIOL, ATPase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OQK
 
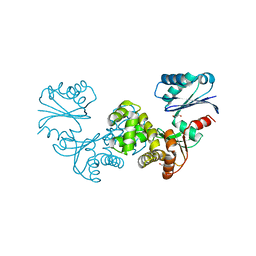 | | Crystal structure of Tannerella forsythia sugar kinase K1058 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine kinase | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
