8YK5
 
 | |
2H9R
 
 | | Docking and dimerization domain (D/D) of the regulatory subunit of the Type II-alpha cAMP-dependent protein kinase A associated with a Peptide derived from an A-kinase anchoring protein (AKAP) | | Descriptor: | 22-mer from A-kinase anchor protein 5, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Newlon, M.G, Roy, M, Morikis, D, Hausken, Z.E, Coghlan, V, Scott, J.D, Jennings, P.A. | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes.
Embo J., 20, 2001
|
|
2DRN
 
 | | Docking and dimerization domain (D/D) of the Type II-alpha regulatory subunity of protein kinase A (PKA) in complex with a peptide from an A-kinase anchoring protein | | Descriptor: | 24-residues peptide from an a-kinase anchoring protein, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Newlon, M.G, Roy, M, Morikis, D, Hausken, Z.E, Coghlan, V, Scott, J.D, Jennings, P.A. | | Deposit date: | 2006-06-11 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes.
Embo J., 20, 2001
|
|
1WTF
 
 | | Crystal structure of Bacillus thermoproteolyticus Ferredoxin Variants Containing Unexpected [3Fe-4S] Cluster that is linked to Coenzyme A at 1.6 A Resolution | | Descriptor: | COENZYME A, FE3-S4 CLUSTER, Ferredoxin, ... | | Authors: | Shirakawa, T, Takahashi, Y, Wada, K, Hirota, J, Takao, T, Ohmori, D, Fukuyama, K. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of variant molecules of Bacillus thermoproteolyticus ferredoxin: crystal structure reveals bound coenzyme A and an unexpected [3Fe-4S] cluster associated with a canonical [4Fe-4S] ligand motif
Biochemistry, 44, 2005
|
|
1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
1YVZ
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[(2,4-DICHLOROBENZOYL)(ISOPROPYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
1YUY
 
 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE GENOTYPE 2a | | Descriptor: | RNA-Dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
2MGW
 
 | | Solution Structure of the UBA Domain of Human NBR1 | | Descriptor: | Next to BRCA1 gene 1 protein | | Authors: | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2013-11-09 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
2MJ5
 
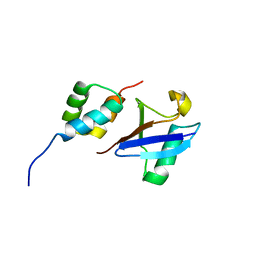 | | Structure of the UBA Domain of Human NBR1 in Complex with Ubiquitin | | Descriptor: | Next to BRCA1 gene 1 protein, Polyubiquitin-C | | Authors: | Walinda, E, Morimoto, D, Sugase, K, Komatsu, M, Tochio, H, Shirakawa, M. | | Deposit date: | 2013-12-25 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin.
J.Biol.Chem., 289, 2014
|
|
2D3U
 
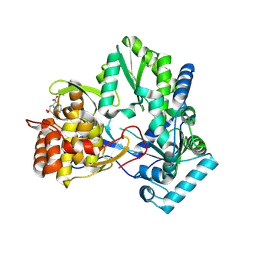 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D41
 
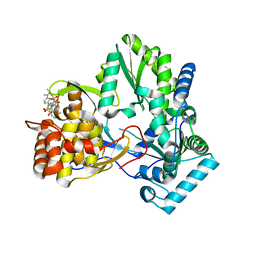 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside inhibitor | | Descriptor: | 5'-ACETYL-4-{[(2,4-DIMETHYLPHENYL)SULFONYL]AMINO}-2,2'-BITHIOPHENE-5-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-05 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3Z
 
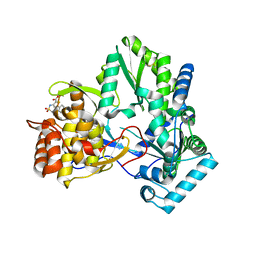 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
1UER
 
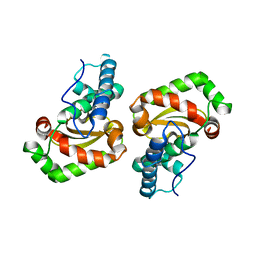 | | Crystal structure of Porphyromonas gingivalis SOD | | Descriptor: | FE (III) ION, superoxide dismutase | | Authors: | Yamakura, F, Sugio, S, Hiraoka, B.Y, Yokota, T, Ohmori, D. | | Deposit date: | 2003-05-20 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pronounced conversion of the metal-specific activity of superoxide dismutase from Porphyromonas gingivalis by the mutation of a single amino acid (Gly155Thr) located apart from the active site
Biochemistry, 42, 2003
|
|
1UES
 
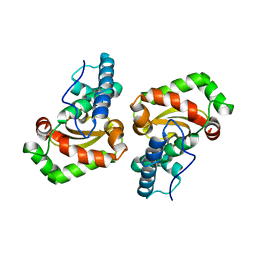 | | Crystal structure of Porphyromonas gingivalis SOD | | Descriptor: | MANGANESE (II) ION, superoxide dismutase | | Authors: | Yamakura, F, Sugio, S, Hiraoka, B.Y, Yokota, T, Ohmori, D. | | Deposit date: | 2003-05-20 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pronounced conversion of the metal-specific activity of superoxide dismutase from Porphyromonas gingivalis by the mutation of a single amino acid (Gly155Thr) located apart from the active site
Biochemistry, 42, 2003
|
|
3B0F
 
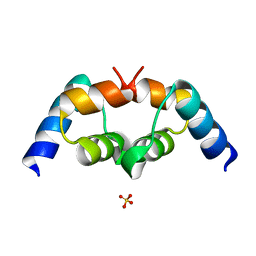 | | Crystal structure of the UBA domain of p62 and its interaction with ubiquitin | | Descriptor: | SULFATE ION, Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
1R2A
 
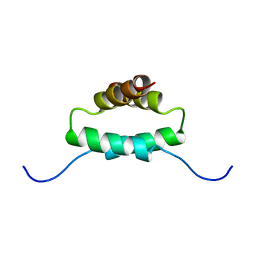 | | THE MOLECULAR BASIS FOR PROTEIN KINASE A ANCHORING REVEALED BY SOLUTION NMR | | Descriptor: | PROTEIN (CAMP-DEPENDENT PROTEIN KINASE TYPE II REGULATORY SUBUNIT) | | Authors: | Newlon, M.G, Roy, M, Morikis, D, Hausken, Z.E, Coghlan, V, Scott, J.D, Jennings, P.A. | | Deposit date: | 1998-12-07 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The molecular basis for protein kinase A anchoring revealed by solution NMR.
Nat.Struct.Biol., 6, 1999
|
|
7CNE
 
 | |
7CJV
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a dilute environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
7CJW
 
 | | Solution structure of monomeric superoxide dismutase 1 with an additional mutation H46W in a crowded environment | | Descriptor: | Monomeric Human Cu,Zn Superoxide dismutase | | Authors: | Iwakawa, N, Morimoto, D, Walinda, E, Danielsson, J, Shirakawa, M, Sugase, K. | | Deposit date: | 2020-07-14 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Transient Diffusive Interactions with a Protein Crowder Affect Aggregation Processes of Superoxide Dismutase 1 beta-Barrel.
J.Phys.Chem.B, 125, 2021
|
|
7YM0
 
 | | Lysoplasmalogen-specific phospholipase D (LyPls-PLD) with Ca2+ | | Descriptor: | CALCIUM ION, Lysoplasmalogenase | | Authors: | Yasutake, Y, Sakasegawa, S, Sugimori, D, Murayama, K. | | Deposit date: | 2022-07-27 | | Release date: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
4HYQ
 
 | |
7D8K
 
 | | Solution structure of the methyl-CpG binding domain of MBD6 from Arabidopsis thaliana | | Descriptor: | Methyl-CpG-binding domain-containing protein 6 | | Authors: | Mahana, Y, Ohki, I, Walinda, E, Morimoto, D, Sugase, K, Shirakawa, M. | | Deposit date: | 2020-10-08 | | Release date: | 2021-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Methylated DNA Recognition by the Methyl-CpG Binding Domain of MBD6 from Arabidopsis thaliana .
Acs Omega, 7, 2022
|
|
7S1G
 
 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1H
 
 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
