7BJV
 
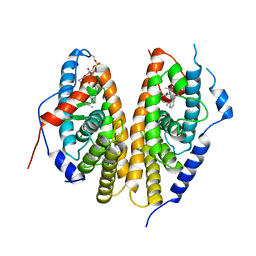 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI09181 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
7BJU
 
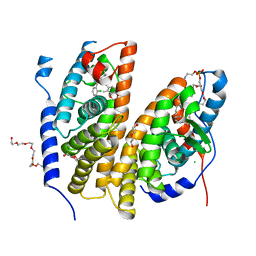 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI08346 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
4DGK
 
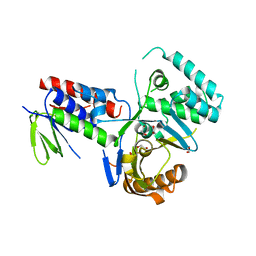 | | Crystal structure of Phytoene desaturase CRTI from Pantoea ananatis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phytoene dehydrogenase | | Authors: | Schaub, P, Yu, Q, Gemmecker, S, Poussin-Courmontagne, P, Mailliot, J, McEwen, A.G, Ghisla, S, Beyer, P, Cavarelli, J. | | Deposit date: | 2012-01-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | On the structure and function of the phytoene desaturase CRTI from Pantoea ananatis, a membrane-peripheral and FAD-dependent oxidase/isomerase.
Plos One, 7, 2012
|
|
5LYV
 
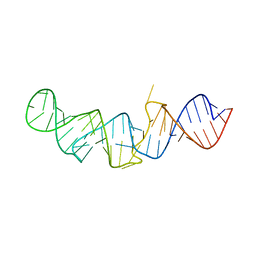 | | The crystal structure of 7SK 5'-hairpin - Osmium derivative | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
5MFT
 
 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-1-bromo-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5MFR
 
 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5MFS
 
 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5LYS
 
 | | The crystal structure of 7SK 5'-hairpin - Gold derivative | | Descriptor: | 7SK RNA, GOLD ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
1OYJ
 
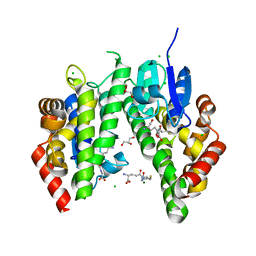 | | Crystal structure solution of Rice GST1 (OsGSTU1) in complex with glutathione. | | Descriptor: | CHLORIDE ION, GLUTATHIONE, GLYCEROL, ... | | Authors: | Dixon, D.P, McEwen, A.G, Lapthorn, A.J, Edwards, R. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Forced evolution of a herbicide detoxifying glutathione transferase.
J.Biol.Chem., 278, 2003
|
|
8CEF
 
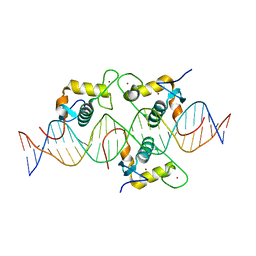 | | Asymmetric Dimerization in a Transcription Factor Superfamily is Promoted by Allosteric Interactions with DNA | | Descriptor: | DNA (26-MER), Nuclear receptor DNA binding domain, ZINC ION | | Authors: | Patel, A.K.M, Shaik, T.B, McEwen, A.G, Moras, D, Klaholz, B.P, Billas, I.M.L. | | Deposit date: | 2023-02-01 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Asymmetric dimerization in a transcription factor superfamily is promoted by allosteric interactions with DNA.
Nucleic Acids Res., 51, 2023
|
|
6SBR
 
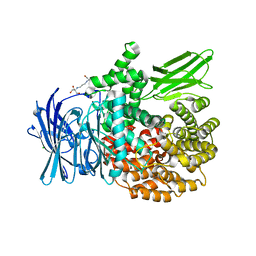 | | The crystal structure of PfA-M1 in complex with 7-amino-1,4-dibromo-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Salomon, E, Schmitt, M, Mouray, E, McEwen, A.G, Torchy, M, Poussin-Courmontagne, P, Alavi, S, Tarnus, C, Cavarelli, J, Florent, I, Albrecht, S. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Aminobenzosuberone derivatives as PfA-M1 inhibitors: Molecular recognition and antiplasmodial evaluation.
Bioorg.Chem., 98, 2020
|
|
6SBQ
 
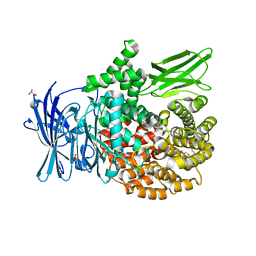 | | The crystal structure of PfA-M1 in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | DI(HYDROXYETHYL)ETHER, M1-family alanyl aminopeptidase, MALONATE ION, ... | | Authors: | Salomon, E, Schmitt, M, Mouray, E, McEwen, A.G, Torchy, M, Poussin-Courmontagne, P, Alavi, S, Tarnus, C, Cavarelli, J, Florent, I, Albrecht, S. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Aminobenzosuberone derivatives as PfA-M1 inhibitors: Molecular recognition and antiplasmodial evaluation.
Bioorg.Chem., 98, 2020
|
|
7Q46
 
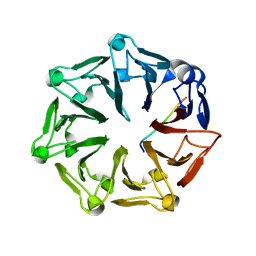 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Pericentriolar material 1 protein | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46002531 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein
To Be Published
|
|
7Q43
 
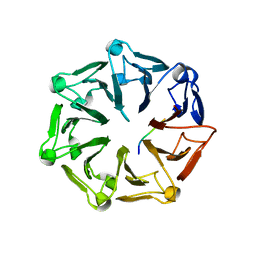 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10 | | Descriptor: | CITRIC ACID, Dedicator of cytokinesis protein 10 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40002346 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10
To Be Published
|
|
7Q41
 
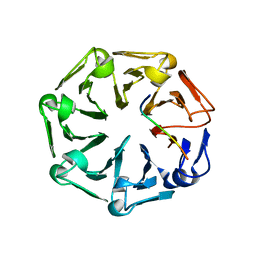 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP) | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Ubiquitin-protein ligase E3A (E6AP) peptide | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.01478052 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP)
To Be Published
|
|
7Q42
 
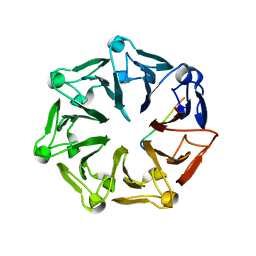 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, CITRIC ACID, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95002484 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B
To Be Published
|
|
7Q45
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1 | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Myelin transcription factor 1 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09999585 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1
To Be Published
|
|
7Q40
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35002232 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2
To Be Published
|
|
7Q44
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35 | | Descriptor: | CITRIC ACID, Deubiquitinase USP35 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.20007777 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35
To Be Published
|
|
7Q71
 
 | | The crystallographic structure of the Ligand Binding domain of the NR7 nuclear receptor from the amphioxus Branchiostoma lanceolatum | | Descriptor: | CHLORIDE ION, Nuclear hormone receptor 7, PHOSPHATE ION | | Authors: | Billas, I.M.L, McEwen, A.G, Hazemann, I, Moras, D, Laudet, V. | | Deposit date: | 2021-11-09 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel nuclear receptor subfamily enlightens the origin of heterodimerization.
Bmc Biol., 20, 2022
|
|
7OON
 
 | | The X-ray structure of heme-bound murine HEBP1 | | Descriptor: | Heme-binding protein 1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Birck, C, Goodfellow, B.J. | | Deposit date: | 2021-05-28 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The SOUL family of heme-binding proteins: Structure and function 15 years later
Coord. Chem. Rev, 448, 2021
|
|
3ZVZ
 
 | | PHD finger of human UHRF1 | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE UHRF1, ZINC ION | | Authors: | Lallous, N, Birck, C, Mc Ewen, A.G, Legrand, P, Samama, J.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | The Phd Finger of Human Uhrf1 Reveals a New Subgroup of Unmethylated Histone H3 Tail Readers.
Plos One, 6, 2011
|
|
3ZVY
 
 | | PHD finger of human UHRF1 in complex with unmodified histone H3 N- terminal tail | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE UHRF1, ... | | Authors: | Lallous, N, Birck, C, Mc Ewen, A.G, Legrand, P, Samama, J.P. | | Deposit date: | 2011-07-28 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phd Finger of Human Uhrf1 Reveals a New Subgroup of Unmethylated Histone H3 Tail Readers.
Plos One, 6, 2011
|
|
5LYU
 
 | | The native crystal structure of 7SK 5'-hairpin | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.C, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
3PY7
 
 | |
