7MJS
 
 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
3TX3
 
 | | CysZ, a putative sulfate permease | | Descriptor: | CHLORIDE ION, LAURYL DIMETHYLAMINE-N-OXIDE, SULFATE ION, ... | | Authors: | Assur, Z, Liu, Q, Hendrickson, W.A, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CysZ, a putative sulfate permease
To be Published
|
|
4Q7C
 
 | | Structure of AF2299, a CDP-alcohol phosphotransferase | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
6UKJ
 
 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | Authors: | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | Deposit date: | 2019-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
6WBX
 
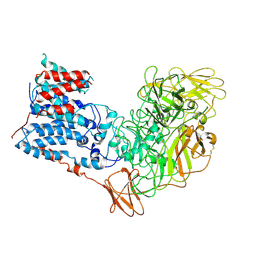 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WBY
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6X0O
 
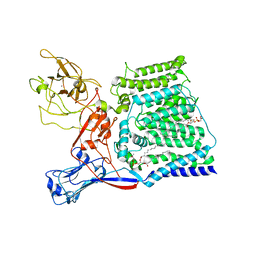 | | Single-Particle Cryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB | | Authors: | Tan, Y.Z, Rodrigues, J, Keener, J.E, Zheng, R.B, Brunton, R, Kloss, B, Giacometti, S.I, Rosario, A.L, Zhang, L, Niederweis, M, Clarke, O.B, Lowary, T.L, Marty, M.T, Archer, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis.
Nat Commun, 11, 2020
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WMV
 
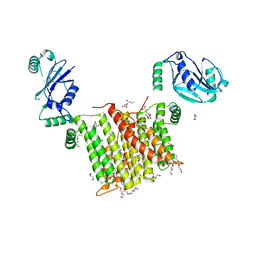 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii with evidence of substrate binding | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,3',3''-phosphanetriyltripropanoic acid, AfCTD-Phosphatidylinositol-phosphate synthase (PIPS) fusion, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F. | | Deposit date: | 2020-04-21 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
5D91
 
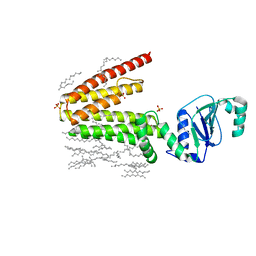 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | Descriptor: | AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, Octadecane, ... | | Authors: | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
5F15
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5EZM
 
 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5EKE
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
5EKP
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (WT) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
5D92
 
 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | Descriptor: | 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, ... | | Authors: | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
8PM2
 
 | | Structure of the murine trace amine-associated receptor TAAR7f bound to N,N-dimethylcyclohexylamine (DMCH) in complex with mini-Gs trimeric G protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gusach, A, Lee, Y, Edwards, P.C, Huang, F, Weyand, S.N, Tate, C.G. | | Deposit date: | 2023-06-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Molecular recognition of an aversive odorant by the murine trace amine-associated receptor TAAR7f.
Biorxiv, 2023
|
|
1NMT
 
 | |
7NMN
 
 | | Rabbit HCN4 stabilised in amphipol A8-35 | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Rabbit HCN4 | | Authors: | Chaves-Sanjuan, A. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
4TKR
 
 | | Native-SAD phasing for ThiT from Listeria monocytogenes serovar. | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, THIAMINE DIPHOSPHATE, Thiamine transporter thia | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKQ
 
 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | Authors: | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8025 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKS
 
 | |
1RLB
 
 | |
