6QGW
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody E6 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoE6, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6HD8
 
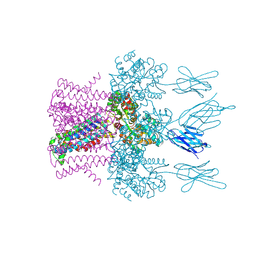 | | Crystal structure of the potassium channel MtTMEM175 in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HD9
 
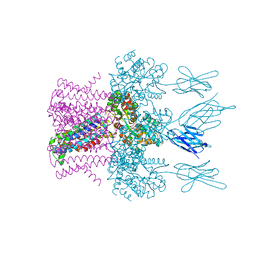 | | Crystal structure of the potassium channel MtTMEM175 with rubidium | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, RUBIDIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
3MEY
 
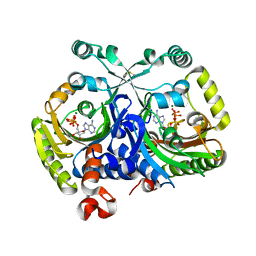 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bll0957 protein, MAGNESIUM ION, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3MF1
 
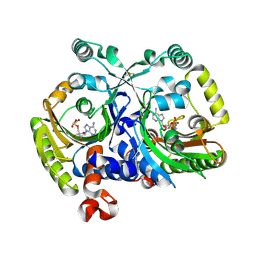 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with an analogue of glycyl adenylate | | Descriptor: | 5'-O-(glycylsulfamoyl)adenosine, Bll0957 protein, ZINC ION | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6HDA
 
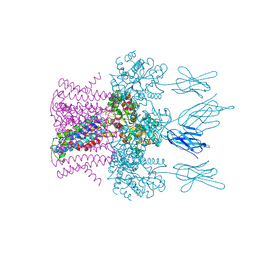 | | Crystal structure of the potassium channel MtTMEM175 with cesium | | Descriptor: | CESIUM ION, DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
7R03
 
 | |
7R04
 
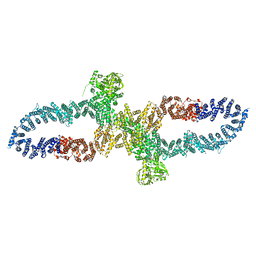 | | Neurofibromin in open conformation | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform I of Neurofibromin | | Authors: | Chaker-Margot, M, Scheffzek, K, Maier, T. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of activation of the tumor suppressor protein neurofibromin.
Mol.Cell, 82, 2022
|
|
6HDC
 
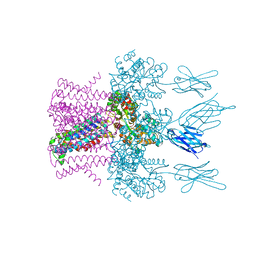 | | Crystal structure of the potassium channel MtTMEM175 T38A variant in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDB
 
 | | Crystal structure of the potassium channel MtTMEM175 with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
3MF2
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Bll0957 protein, GLYCEROL, ... | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8AS3
 
 | | Structure of arrestin2 in complex with 6P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
8AS2
 
 | | Structure of arrestin2 in complex with 4P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
9F42
 
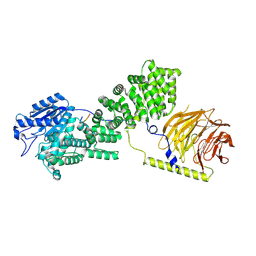 | |
9F43
 
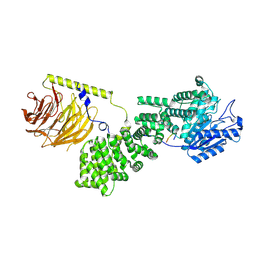 | |
9F44
 
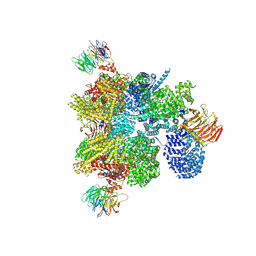 | |
9F45
 
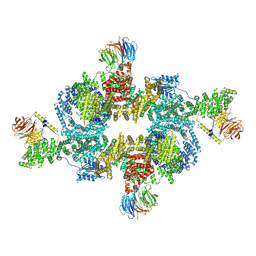 | | cryo-EM structure of human LST2 bound to human mTOR complex 1 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Lateral signaling target protein 2 homolog, Regulatory-associated protein of mTOR, ... | | Authors: | Craigie, L.M, Maier, T. | | Deposit date: | 2024-04-26 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | mTORC1 phosphorylates and stabilizes LST2 to negatively regulate EGFR.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6SWR
 
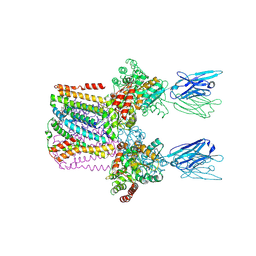 | | Crystal structure of the lysosomal potassium channel MtTMEM175 T38A mutant soaked with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody, Maltose/maltodextrin-binding periplasmic protein,Maltodextrin-binding protein,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2019-09-23 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K + channels.
Elife, 9, 2020
|
|
7R1V
 
 | |
7R1W
 
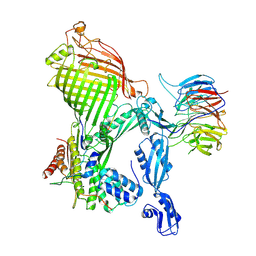 | | E. coli BAM complex (BamABCDE) bound to dynobactin A | | Descriptor: | Dynobactin A, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Hiller, S, Maier, T. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
7NFY
 
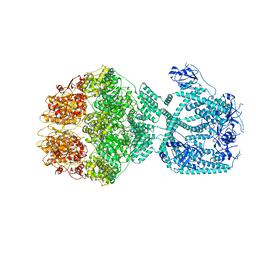 | | P1a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and ATPgS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NG5
 
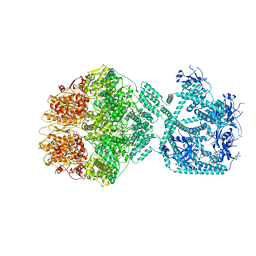 | | P1c-state of wild type human mitochondrial LONP1 protease with bound substrate protein in presence of ATP/ADP mix | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NG4
 
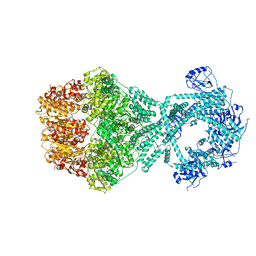 | | P1b-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGC
 
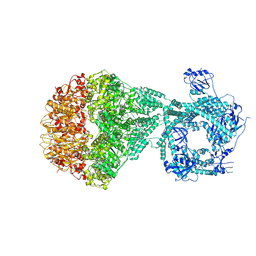 | | P2a-state of wild type human mitochondrial LONP1 protease with bound substrate protein and in presence of ATPgS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NRF
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 2) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
