5T5N
 
 | | Calcium-activated chloride channel bestrophin-1 (BEST1), triple mutant: I76A, F80A, F84A; in complex with an Fab antibody fragment, chloride, and calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fab antibody fragment, ... | | Authors: | Long, S.B, Vaisey, G. | | Deposit date: | 2016-08-31 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct regions that control ion selectivity and calcium-dependent activation in the bestrophin ion channel.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7KR5
 
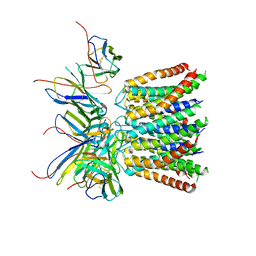 | | Cryo-EM structure of the CRAC channel Orai in an open conformation; H206A gain-of-function mutation in complex with an antibody | | Descriptor: | 19B5 Fab heavy chain, 19B5 Fab light chain, CALCIUM ION, ... | | Authors: | Long, S.B, Hou, X, Outhwaite, I.R. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the calcium release-activated calcium channel Orai in an open conformation.
Elife, 9, 2020
|
|
5VG9
 
 | | Structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase) without a monobody | | Descriptor: | Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
6BBI
 
 | |
6BBH
 
 | |
6BBF
 
 | |
6BBG
 
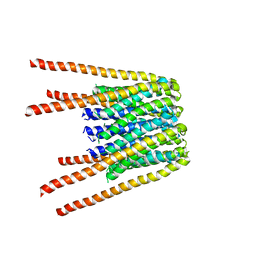 | |
1S64
 
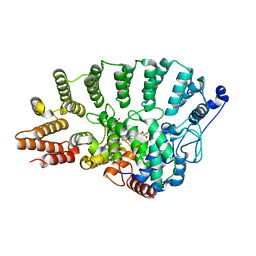 | | Rat protein geranylgeranyltransferase type-I complexed with L-778,123 and a sulfate anion | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[(5-{[4-(3-CHLOROPHENYL)-3-OXOPIPERAZIN-1-YL]METHYL}-1H-IMIDAZOL-1-YL)METHYL]BENZONITRILE, CHLORIDE ION, ... | | Authors: | Reid, T.S, Long, S.B, Beese, L.S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Analysis Reveals that Anticancer Clinical Candidate L-778,123 Inhibits Protein Farnesyltransferase and Geranylgeranyltransferase-I by Different Binding Modes.
Biochemistry, 43, 2004
|
|
6N25
 
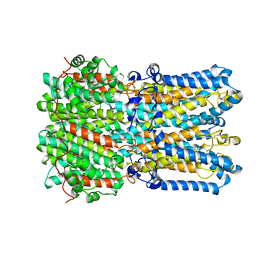 | |
6N27
 
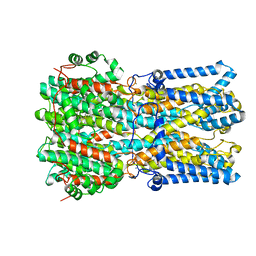 | | BEST1 calcium-bound closed state | | Descriptor: | Bestrophin homolog, CALCIUM ION | | Authors: | Miller, A.N, Vaisey, G, Long, S.B. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanisms of gating in the calcium-activated chloride channel bestrophin.
Elife, 8, 2019
|
|
6N24
 
 | |
6N26
 
 | | BEST1 calcium-free closed state | | Descriptor: | Bestrophin homolog | | Authors: | Miller, A.N, Vaisey, G, Long, S.B. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanisms of gating in the calcium-activated chloride channel bestrophin.
Elife, 8, 2019
|
|
6N23
 
 | |
6N28
 
 | | BEST1 calcium-bound open state | | Descriptor: | Bestrophin homolog, CALCIUM ION | | Authors: | Miller, A.N, Vaisey, G, Long, S.B. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanisms of gating in the calcium-activated chloride channel bestrophin.
Elife, 8, 2019
|
|
4RDQ
 
 | | Calcium-activated chloride channel bestrophin-1, from chicken, in complex with Fab antibody fragments, chloride and calcium | | Descriptor: | 6-cyclohexyl-2-(4-cyclohexylbutyl)-2-({[4-O-(alpha-D-glucopyranosyl)-beta-D-glucopyranosyl]oxy}methyl)hexyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Bestrophin-1, CALCIUM ION, ... | | Authors: | Dickson, V.K, Pedi, L, Long, S.B. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-22 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and insights into the function of a Ca(2+)-activated Cl(-) channel.
Nature, 516, 2014
|
|
3LUT
 
 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Chen, X, Ni, F, Wang, Q, Ma, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
