6PKF
 
 | | Myocilin OLF mutant N428E/D478K | | Descriptor: | GLYCEROL, Myocilin | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PKD
 
 | | Myocilin OLF mutant N428D/D478H | | Descriptor: | Myocilin, SODIUM ION | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4WXU
 
 | | Crystal Structure of the Selenomthionine Incorporated Myocilin Olfactomedin Domain E396D Variant. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
8DAJ
 
 | | Structure and Biochemistry of a Promiscuous Thermophilic Polyhydroxybutyrate Depolymerase from Lihuaxuella thermophilia | | Descriptor: | 1,2-ETHANEDIOL, Esterase, PHB depolymerase family, ... | | Authors: | Thomas, G.M, Quirk, S, Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2022-06-13 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Bioplastic degradation by a polyhydroxybutyrate depolymerase from a thermophilic soil bacterium.
Protein Sci., 31, 2022
|
|
7RTG
 
 | | Crystal Structure of the Human Adenosine Deaminase 1 | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Ma, M.T, Lieberman, R.L, Blazeck, J, Jennings, M.R. | | Deposit date: | 2021-08-13 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Catalytically active holo Homo sapiens adenosine deaminase I adopts a closed conformation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8FRR
 
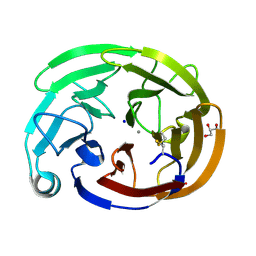 | | Wild-type myocilin olfactomedin domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Ma, M.T, Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2023-01-08 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Competition between inside-out unfolding and pathogenic aggregation in an amyloid-forming beta-propeller.
Nat Commun, 15, 2024
|
|
8DOT
 
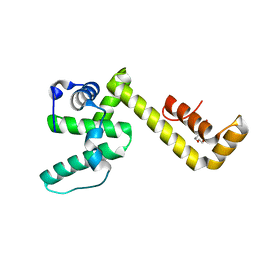 | |
7T8D
 
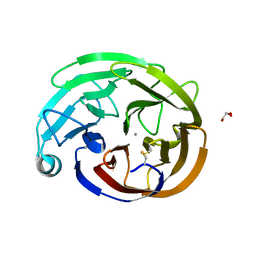 | | Myocilin OLF mutant V449I | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-12-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
5HQ3
 
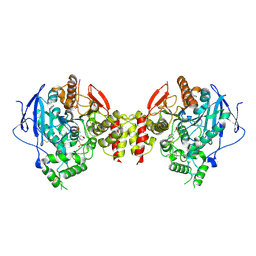 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
7SIJ
 
 | | Myocilin OLF mutant E352K | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SJV
 
 | | Myocilin OLF mutant T353I | | Descriptor: | CALCIUM ION, Myocilin, C-terminal fragment, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SKF
 
 | | Myocilin OLF mutant A445V | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.282 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SJT
 
 | | Myocilin OLF mutant K398R | | Descriptor: | CALCIUM ION, Myocilin, C-terminal fragment, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SKE
 
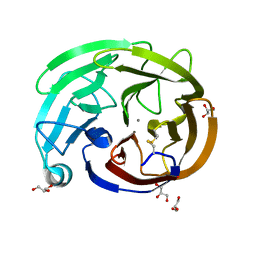 | | Myocilin OLF mutant R296H | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SJU
 
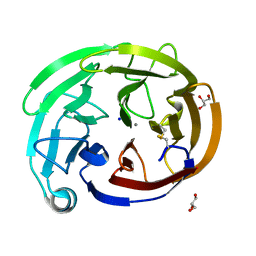 | | Myocilin OLF mutant T293K | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SIB
 
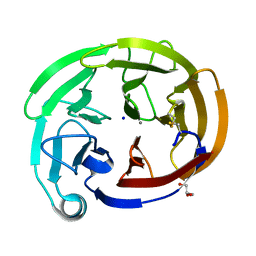 | | Myocilin OLF mutant K500R | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SKD
 
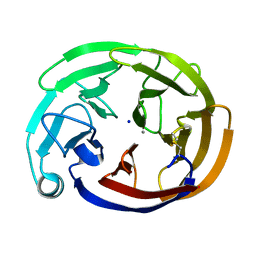 | | Myocilin OLF mutant S331L | | Descriptor: | CALCIUM ION, Myocilin, C-terminal fragment, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SJW
 
 | | Myocilin OLF mutant L303I | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-19 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
7SKG
 
 | | Myocilin OLF mutant V329M | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Scelsi, H.S, Barlow, B.M, Lieberman, R.L. | | Deposit date: | 2021-10-20 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Quantitative differentiation of benign and misfolded glaucoma-causing myocilin variants on the basis of protein thermal stability.
Dis Model Mech, 16, 2023
|
|
6MOZ
 
 | | Structure of acid-beta-glucosidase in complex with an aromatic pyrrolidine iminosugar inhibitor | | Descriptor: | (2R,3S,4R)-2-{[4-(3,5-dichlorophenyl)-1H-1,2,3-triazol-1-yl]methyl}pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Patterson-Orazem, A.C, Lieberman, R.L. | | Deposit date: | 2018-10-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring substituent diversity on pyrrolidine-aryltriazole iminosugars: Structural basis of beta-glucocerebrosidase inhibition.
Bioorg.Chem., 86, 2019
|
|
6NAX
 
 | | Olfactomedin domain of mouse myocilin | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Patterson-Orazem, A.C, Hill, S.E, Lieberman, R.L. | | Deposit date: | 2018-12-06 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Differential Misfolding Properties of Glaucoma-Associated Olfactomedin Domains from Humans and Mice.
Biochemistry, 58, 2019
|
|
3RIK
 
 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | Descriptor: | (3S,4R,5R,6S)-1-(2-hydroxyethyl)azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Orwig, S.D, Lieberman, R.L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
8DYK
 
 | |
3RIL
 
 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | Descriptor: | (3S,4R,5R,6S)-azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Orwig, S.D, Lieberman, R.L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
6ASP
 
 | | Structure of Grp94 with methyl 3-chloro-2-(2-(1-(2-ethoxybenzyl)-1 H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, a Grp94-selective inhibitor and promising therapeutic lead for treating myocilin-associated glaucoma | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endoplasmin, GLYCEROL, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
