2PY9
 
 | |
2C7U
 
 | | Conflicting selective forces affect CD8 T-cell receptor contact sites in an HLA-A2 immunodominant HIV epitope. | | Descriptor: | BETA-2-MICROGLOBULIN, GAG PROTEIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Iversen, A.K, Stewart-Jones, G, Learn, G.H, Christie, N, Sylvester-Hviid, C, Armitage, A.E, Kaul, R, Beattie, T, Lee, J.K, Li, Y, Chotiyarnwong, P, Dong, T, Xu, X, Luscher, M.A, MacDonald, K, Ullum, H, Klarlund-Pedersen, B, Skinhoj, P, Fugger, J.L, Buus, S, Mullins, J.I, Jones, E.Y, van der Merwe, P.A, McMichael, A.J. | | Deposit date: | 2005-11-29 | | Release date: | 2006-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Conflicting Selective Forces Affect T Cell Receptor Contacts in an Immunodominant Human Immunodeficiency Virus Epitope.
Nat.Immunol., 7, 2006
|
|
2AXY
 
 | | Crystal Structure of KH1 domain of human Poly(C)-binding protein-2 with C-rich strand of human telomeric DNA | | Descriptor: | C-rich strand of human telomeric dna, Poly(rC)-binding protein 2 | | Authors: | Du, Z, Lee, J.K, Tjhen, R.J, Li, S, Stroud, R.M, James, T.L. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the First KH Domain of Human Poly(C)-binding Protein-2 in Complex with a C-rich Strand of Human Telomeric DNA at 1.7 A
J.Biol.Chem., 280, 2005
|
|
1J4N
 
 | | Crystal Structure of the AQP1 water channel | | Descriptor: | AQUAPORIN 1, nonyl beta-D-glucopyranoside | | Authors: | Sui, H, Han, B.-G, Lee, J.K, Walian, P, Jap, B.K. | | Deposit date: | 2001-10-19 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of water-specific transport through the AQP1 water channel.
Nature, 414, 2001
|
|
5W8X
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations-Bound to UDP | | Descriptor: | Lipid-A-disaccharide synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
5W8N
 
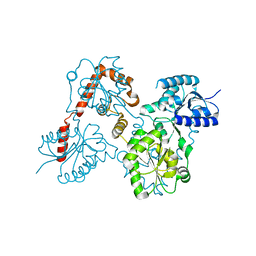 | |
5W8S
 
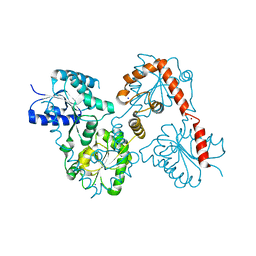 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations | | Descriptor: | LITHIUM ION, Lipid-A-disaccharide synthase, SODIUM ION | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
4YJ0
 
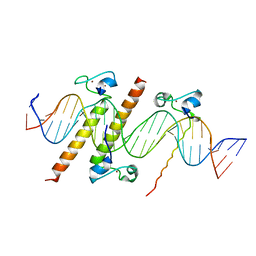 | | Crystal structure of the DM domain of human DMRT1 bound to 25mer target DNA | | Descriptor: | DNA (25-MER), Doublesex- and mab-3-related transcription factor 1, ZINC ION | | Authors: | Murphy, M.W, Lee, J.K, Rojo, S, Gearhart, M.D, Kurahashi, K, Banerjee, S, Loeuille, G, Bashamboo, A, McElreavey, K, Zarkower, D, Aihara, H, Bardwell, V.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.814 Å) | | Cite: | An ancient protein-DNA interaction underlying metazoan sex determination.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4J6E
 
 | | Structure of LPXI D225A Mutant | | Descriptor: | (2R,3R,4R,5S,6R)-2-{[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)-3-{[(3R)-3-hydroxytetradecanoyl]amino}tetrahydro-2H-pyran-4-yl (3R)-3-hydroxytetradecanoate, UDP-2,3-diacylglucosamine pyrophosphatase LpxI | | Authors: | Metzger IV, L.E, Lee, J.K, Finer-Moore, J.S, Raetz, C.R.H, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | LpxI structures reveal how a lipid A precursor is synthesized.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4GGM
 
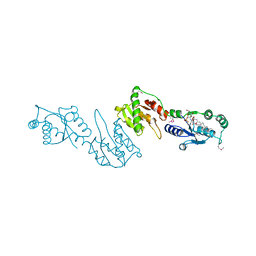 | | Structure of LpxI | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, MAGNESIUM ION, UDP-2,3-diacylglucosamine pyrophosphatase LpxI | | Authors: | Metzger IV, L.E, Lee, J.K, Finer-Moore, J.S, Raetz, C.R.H, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | LpxI structures reveal how a lipid A precursor is synthesized.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3P14
 
 | |
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNF
 
 | | Cryo-EM structure of RNase-untreated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNI
 
 | | Cryo-EM structure of RNase-treated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNK
 
 | | Cryo-EM structure of RNase-untreated RESC-B in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 10 (RESC10), RNA-editing substrate-binding complex protein 11 (RESC11), RNA-editing substrate-binding complex protein 13 (RESC13), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8F3D
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, beta-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase alpha-subunit, 3-methylcrotonyl-CoA carboxylase beta-subunit, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
8F41
 
 | | 3-methylcrotonyl-CoA carboxylase in filament, alpha-subunit centered | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, alpha-subunit, beta-subunit, ... | | Authors: | Hu, J.J, Lee, J.K.J, Liu, Y.T, Yu, C, Huang, L, Afasizheva, I, Afasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-11-10 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Discovery, structure, and function of filamentous 3-methylcrotonyl-CoA carboxylase.
Structure, 31, 2023
|
|
4TQ1
 
 | | Crystal structure of human ATG5-TECAIR | | Descriptor: | Autophagy protein 5, Tectonin beta-propeller repeat-containing protein 1 | | Authors: | Kim, J.H, Hong, S.B, Song, H.K. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners
Autophagy, 11, 2015
|
|
4TQ0
 
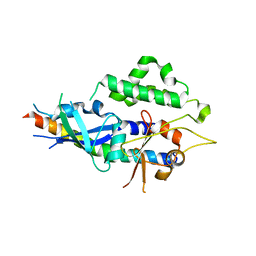 | | Crystal structure of human ATG5-ATG16N69 | | Descriptor: | Autophagy protein 5, Autophagy-related protein 16-1 | | Authors: | Kim, J.H, Hong, S.B, Song, H.K. | | Deposit date: | 2014-06-10 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners
Autophagy, 11, 2015
|
|
1AVU
 
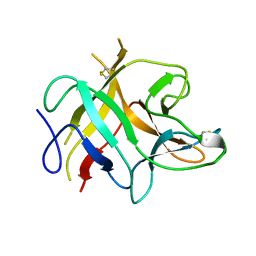 | | TRYPSIN INHIBITOR FROM SOYBEAN (STI) | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-20 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
2ORY
 
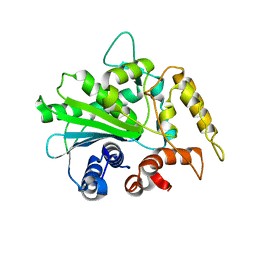 | | Crystal structure of M37 lipase | | Descriptor: | Lipase | | Authors: | Jung, S.K, Jeong, D.G, Kim, S.J. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum.
Proteins, 71, 2008
|
|
1AVW
 
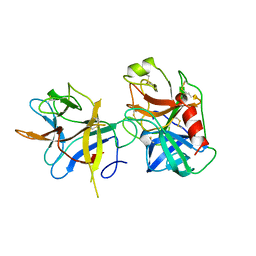 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
1AVX
 
 | | COMPLEX PORCINE PANCREATIC TRYPSIN/SOYBEAN TRYPSIN INHIBITOR, TETRAGONAL CRYSTAL FORM | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR | | Authors: | Song, H.K, Suh, S.W. | | Deposit date: | 1997-09-21 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kunitz-type soybean trypsin inhibitor revisited: refined structure of its complex with porcine trypsin reveals an insight into the interaction between a homologous inhibitor from Erythrina caffra and tissue-type plasminogen activator.
J.Mol.Biol., 275, 1998
|
|
