6WB5
 
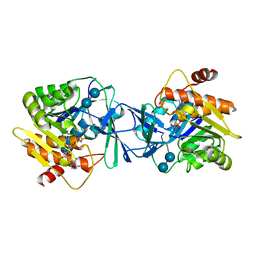 | | Microbiome-derived Acarbose Kinase Mak1 as a Complex with Acarbose and AMP-PNP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose Kinase Mak1, MANGANESE (II) ION, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
6WB7
 
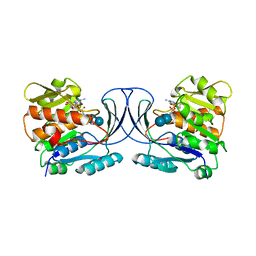 | | Acarbose Kinase AcbK as a Complex with Acarbose and AMP-PNP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose 7(IV)-phosphotransferase, MANGANESE (II) ION, ... | | Authors: | Jeffrey, P.D, Balaich, J.N, Estrella, M.A, Donia, M.S. | | Deposit date: | 2020-03-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The human microbiome encodes resistance to the antidiabetic drug acarbose.
Nature, 600, 2021
|
|
6O0W
 
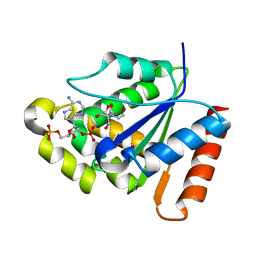 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0V
 
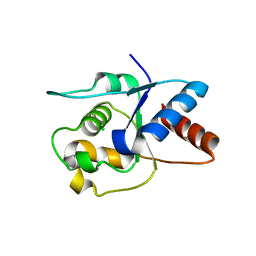 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0S
 
 | | Crystal structure of the tandem SAM domains from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0T
 
 | | Crystal structure of selenomethionine labelled tandem SAM domains (L446M:L505M:L523M mutant) from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O1B
 
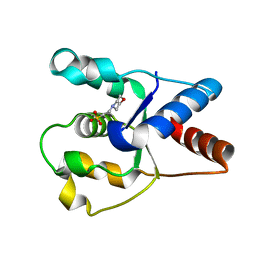 | | Crystal structure of the TIR domain G601P mutant from human SARM1, crystal form 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-18 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0R
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with glycerol | | Descriptor: | GLYCEROL, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0U
 
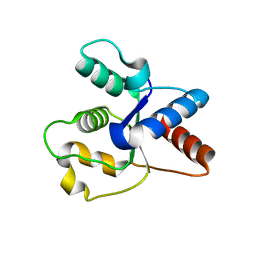 | | Crystal structure of the TIR domain H685A mutant from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0Q
 
 | | Crystal structure of the TIR domain from human SARM1 in complex with ribose | | Descriptor: | CHLORIDE ION, Sterile alpha and TIR motif-containing protein 1, beta-D-ribofuranose | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
5XP6
 
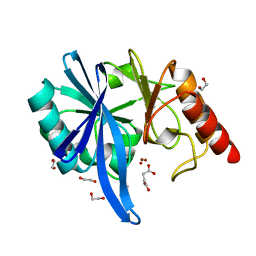 | | native structure of NDM-1 crystallized at pH5.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G, Lai, J, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
6GCJ
 
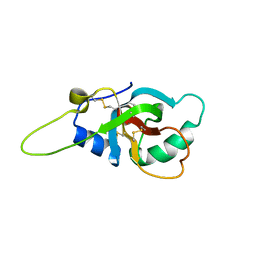 | | Solution structure of the RodA hydrophobin from Aspergillus fumigatus | | Descriptor: | Hydrophobin | | Authors: | Pille, A, Kwan, A, Aimanianda, V, Latge, J.-P, Sunde, M, Guijarro, J.I. | | Deposit date: | 2018-04-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-09-04 | | Method: | SOLUTION NMR | | Cite: | Assembly and disassembly of Aspergillus fumigatus conidial rodlets
Cell Surf, 2019
|
|
1JPS
 
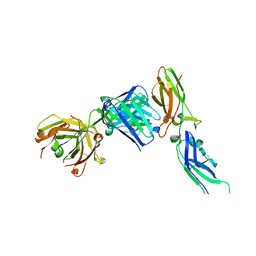 | | Crystal structure of tissue factor in complex with humanized Fab D3h44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, light chain, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
1JPT
 
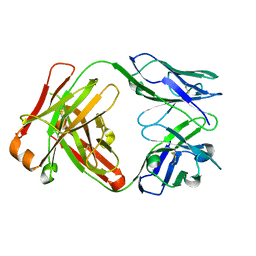 | | Crystal Structure of Fab D3H44 | | Descriptor: | immunoglobulin Fab D3H44, heavy chain, immunoglobulin Fab D3h44, ... | | Authors: | Faelber, K, Kirchhofer, D, Presta, L, Kelley, R.F, Muller, Y.A. | | Deposit date: | 2001-08-03 | | Release date: | 2002-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A resolution crystal structures of tissue factor in complex with humanized Fab D3h44 and of free humanized Fab D3h44: revisiting the solvation of antigen combining sites.
J.Mol.Biol., 313, 2001
|
|
5K5B
 
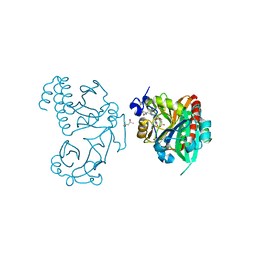 | | Wild-type PAS-GAF fragment from Deinococcus radiodurans BphP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, ... | | Authors: | Takala, H, Edlund, P, Claesson, E, Ihalainen, J.A, Westenhoff, S. | | Deposit date: | 2016-05-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography.
Sci Rep, 6, 2016
|
|
3KE1
 
 | |
8RUS
 
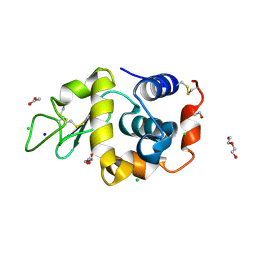 | | Hen egg-white lysozyme (HEWL) structure from EuXFEL FXE, multi-hit Droplet-on-Demand (DoD) injection, 9.3 keV photon energy, space group P432121 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Perrett, S, van Thor, J.J. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kilohertz droplet-on-demand serial femtosecond crystallography at the European XFEL station FXE.
Struct Dyn., 11, 2024
|
|
6TRK
 
 | | Phl p 6 fold stabilized mutant - S46Y | | Descriptor: | Pollen allergen Phl p 6, ZINC ION | | Authors: | Soh, W.T, Brandstetter, H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In silico Design of Phl p 6 Variants With Altered Fold-Stability Significantly Impacts Antigen Processing, Immunogenicity and Immune Polarization.
Front Immunol, 11, 2020
|
|
6YPB
 
 | | NUDIX1 hydrolase from Rosa x hybrida | | Descriptor: | Geranyl diphosphate phosphohydrolase | | Authors: | Degut, C, Rety, S, Tisne, C, Baudino, S. | | Deposit date: | 2020-04-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional diversification in the Nudix hydrolase gene family drives sesquiterpene biosynthesis in Rosa × wichurana.
Plant J., 104, 2020
|
|
6YPF
 
 | | NUDIX1 hydrolase from Rosa x hybrida in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate phosphohydrolase | | Authors: | Degut, C, Rety, S, Tisne, C, Baudino, S. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Functional diversification in the Nudix hydrolase gene family drives sesquiterpene biosynthesis in Rosa × wichurana.
Plant J., 104, 2020
|
|
6OKF
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
6OLT
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C12-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
6OKC
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C12-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
6OKG
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C16-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, N-[2-(hexadecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
7JVM
 
 | |
