6O8Z
 
 | |
6WUB
 
 | |
6WUA
 
 | |
6WU9
 
 | |
6W6P
 
 | |
6O8X
 
 | |
6O8W
 
 | |
6O8Y
 
 | |
3IYI
 
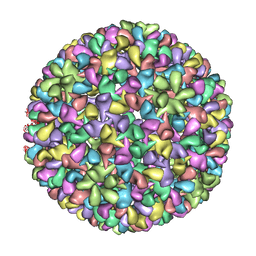 | | P22 expanded head coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | P22 coat protein in procapsid shells | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
3IYH
 
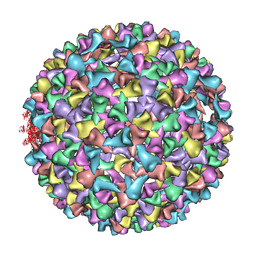 | | P22 procapsid coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical cross-links | | Descriptor: | Coat protein | | Authors: | Parent, K.N, Khayat, R, Tu, L.H, Suhanovsky, M.M, Cortines, J.R, Teschke, C.M, Johnson, J.E, Baker, T.S. | | Deposit date: | 2009-12-14 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | P22 coat protein structures reveal a novel mechanism for capsid maturation: stability without auxiliary proteins or chemical crosslinks
Structure, 18, 2010
|
|
3QPR
 
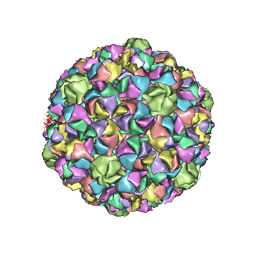 | | HK97 Prohead I encapsidating inactive virally encoded protease | | Descriptor: | Major capsid protein | | Authors: | Huang, R.K, Khayat, R, Lee, K.K, Gertsman, I, Duda, R.L, Hendrix, R.W, Johnson, J.E. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | The Prohead-I structure of bacteriophage HK97: implications for scaffold-mediated control of particle assembly and maturation.
J.Mol.Biol., 408, 2011
|
|
1JQ7
 
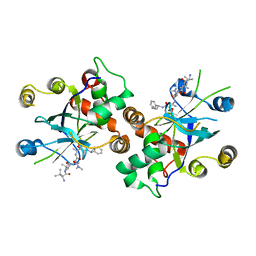 | | HCMV protease dimer-interface mutant, S225Y complexed to Inhibitor BILC 408 | | Descriptor: | ASSEMBLIN, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Batra, R, Khayat, R, Tong, L. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease.
Nat.Struct.Biol., 8, 2001
|
|
1JQ6
 
 | |
1LXJ
 
 | | X-RAY STRUCTURE OF YBL001c NORTHEAST STRUCTURAL GENOMICS (NESG) CONSORTIUM TARGET YTYst72 | | Descriptor: | HYPOTHETICAL 11.5KDA PROTEIN IN HTB2-NTH2 INTERGENIC REGION, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURES OF MTH1187 AND ITS YEAST ORTHOLOG YBL001C
Proteins, 52, 2003
|
|
1LXN
 
 | | X-RAY STRUCTURE OF MTH1187 NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT272 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1187, SULFATE ION | | Authors: | Tao, X, Khayat, R, Christendat, D, Savchenko, A, Xu, X, Edwards, A, Arrowsmith, C.H, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-06-05 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of MTH1187 and its Yeast Ortholog YBL001C
Proteins, 52, 2003
|
|
4JY4
 
 | | Crystal structure of human Fab PGT121, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PGT121 heavy chain, ... | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
4JY6
 
 | | Crystal structure of human Fab PGT123, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGT123 heavy chain, PGT123 light chain, ... | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans.
Plos Pathog., 9, 2013
|
|
4JY5
 
 | | Crystal structure of human Fab PGT122, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | GLYCEROL, PGT122 heavy chain, PGT122 light chain | | Authors: | Julien, J.-P, Diwanji, D.C, Burton, D.R, Wilson, I.A. | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans.
Plos Pathog., 9, 2013
|
|
4JM2
 
 | |
4JM4
 
 | | Crystal Structure of PGT 135 Fab | | Descriptor: | PGT 135 Heavy Chain, PGT 135 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-29 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4FQM
 
 | | Structure of B/Brisbane/60/2008 Influenza Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQL
 
 | | Influenza B HA Antibody (Fab) CR8033 | | Descriptor: | monoclonal antibody CR8033 heavy chain, monoclonal antibody CR8033 light chain | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQV
 
 | | Crystal structure of broadly neutralizing antibody CR9114 bound to H7 influenza hemagglutinin | | Descriptor: | Antibody CR9114 heavy chain, Antibody CR9114 light chain, Hemagglutinin HA1, ... | | Authors: | Ekiert, D.C, Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQH
 
 | | Crystal Structure of Fab CR9114 | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, antibody CR9114 heavy chain, ... | | Authors: | Dreyfus, C, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
4FQK
 
 | | Influenza B/Brisbane/60/2008 hemagglutinin Fab CR8059 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody CR8059 Heavy Chain, ... | | Authors: | Dreyfus, C, Laursen, N.S, Wilson, I.A. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (5.65 Å) | | Cite: | Highly conserved protective epitopes on influenza B viruses.
Science, 337, 2012
|
|
