3EYK
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.J, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3EYM
 
 | | Structure of Influenza Haemagglutinin in complex with an inhibitor of membrane fusion | | Descriptor: | 2-tert-butylbenzene-1,4-diol, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Russell, R.J, Kerry, P.S, Stevens, D.A, Steinhauer, D.A, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2008-10-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3L4Q
 
 | | Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein | | Descriptor: | GLYCEROL, Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Hale, B.G, Kerry, P.S, Jackson, D, Precious, B.L, Gray, A, Killip, M.J, Randall, R.E, Russell, R.J. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3O9K
 
 | | Influenza NA in complex with compound 6 | | Descriptor: | 5-acetamido-2,6-anhydro-3,5-dideoxy-3-[(2E)-3-(4-methylphenyl)prop-2-en-1-yl]-D-glycero-D-galacto-non-2-enonic acid, Neuraminidase | | Authors: | Russell, R.J, Kerry, P.S. | | Deposit date: | 2010-08-04 | | Release date: | 2010-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4945 Å) | | Cite: | Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase.
Nat Commun, 1, 2010
|
|
3O9J
 
 | | Influenza NA in complex with compound 5 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 5-acetamido-2,6-anhydro-3,5-dideoxy-3-prop-2-en-1-yl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Russell, R.J, Kerry, P.S. | | Deposit date: | 2010-08-04 | | Release date: | 2010-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.0002 Å) | | Cite: | Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase.
Nat Commun, 1, 2010
|
|
7NAI
 
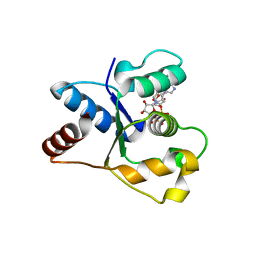 | | Crystal structure of the TIR domain from human SARM1 in complex with 3AD | | Descriptor: | Sterile alpha and TIR motif-containing protein 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAJ
 
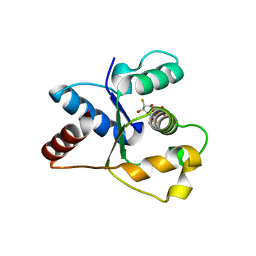 | | Crystal structure of the TIR domain from human SARM1 in complex with ara-2'F-ADPR | | Descriptor: | 1,4-anhydro-2-deoxy-2-fluoro-5-O-[(S)-hydroxy(phosphonooxy)phosphoryl]-D-arabinitol, Sterile alpha and TIR motif-containing protein 1 | | Authors: | Shi, Y, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAG
 
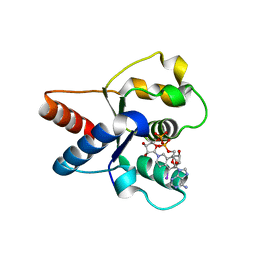 | | Crystal structure of the TIR domain from human SARM1 in complex with 1AD | | Descriptor: | Sterile alpha and TIR motif-containing protein 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Bosanac, T, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAH
 
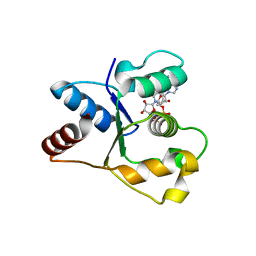 | | Crystal structure of the TIR domain from human SARM1 in complex with 2AD | | Descriptor: | Sterile alpha and TIR motif-containing protein 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-(8-oxidanylidene-7~{H}-2,7-naphthyridin-2-yl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCY
 
 | | Crystal structure of the ligand-free ARM domain from Drosophila SARM1 | | Descriptor: | Isoform B of NAD(+) hydrolase sarm1 | | Authors: | Gu, W, Nanson, J.D, Luo, Z, McGuinness, H.Y, Manik, M.K, Jia, X, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7LCZ
 
 | | Crystal structure of the ARM domain from Drosophila SARM1 in complex with NMN | | Descriptor: | 1,2-ETHANEDIOL, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Isoform B of NAD(+) hydrolase sarm1, ... | | Authors: | Gu, W, Nanson, J.D, Luo, Z, Jia, X, Manik, M.K, Ve, T, Kobe, B. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
