1OKT
 
 | | X-ray Structure of Glutathione S-Transferase from the Malarial Parasite Plasmodium falciparum | | Descriptor: | FORMIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Fritz-Wolf, K, Becker, A, Rahlfs, s, Harwaldt, P, Schirmer, R.H, Kabsch, W, Becker, K. | | Deposit date: | 2003-07-29 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of Glutathione S-Transferase from the Malarial Parasite Plasmodium Falciparum
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1GK1
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
1GK0
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
2PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI | | Descriptor: | CHLORIDE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-26 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
1G6T
 
 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1G6S
 
 | | STRUCTURE OF EPSP SYNTHASE LIGANDED WITH SHIKIMATE-3-PHOSPHATE AND GLYPHOSATE | | Descriptor: | EPSP SYNTHASE, FORMIC ACID, GLYPHOSATE, ... | | Authors: | Schonbrunn, E, Eschenburg, S, Shuttleworth, W, Schloss, J.V, Amrhein, N, Evans, J.N.S, Kabsch, W. | | Deposit date: | 2000-11-07 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1H18
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate | | Descriptor: | FORMATE ACETYLTRANSFERASE 1, L-TREITOL, PYRUVIC ACID, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1H17
 
 | |
1H16
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate and CoA | | Descriptor: | COENZYME A, FORMATE ACETYLTRANSFERASE 1, L-TREITOL, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1ICJ
 
 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1CRK
 
 | | MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Schnyder, T, Wallimann, T, Kabsch, W. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mitochondrial creatine kinase.
Nature, 381, 1996
|
|
1CM5
 
 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | Descriptor: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
4RLM
 
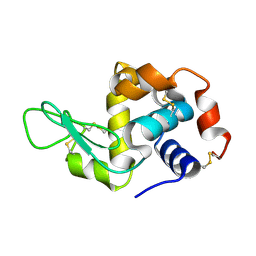 | | Hen egg-white lysozyme solved from serial crystallography at a synchrotron source, data processed with CrystFEL | | Descriptor: | Lysozyme C | | Authors: | Botha, S, Nass, K, Barends, T, Kabsch, W, Latz, B, Dworkowski, F, Foucar, L, Panepucci, E, Wang, M, Shoeman, R, Schlichting, I, Doak, R.B. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RLN
 
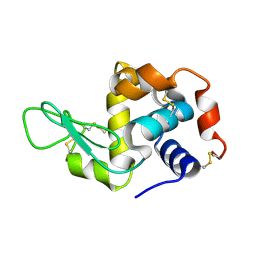 | | Hen egg-white lysozyme solved from serial crystallography at a synchrotron source, data processed with nXDS | | Descriptor: | Lysozyme C | | Authors: | Botha, S, Nass, K, Barends, T, Kabsch, W, Latz, B, Dworkowski, F, Foucar, L, Panepucci, E, Wang, M, Shoeman, R, Schlichting, I, Doak, R.B. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3PFL
 
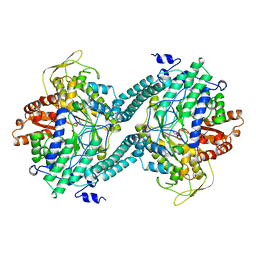 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI IN COMPLEX WITH SUBSTRATE ANALOGUE OXAMATE | | Descriptor: | OXAMIC ACID, PROTEIN (FORMATE ACETYLTRANSFERASE 1) | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
821P
 
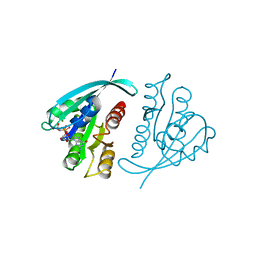 | | THREE-DIMENSIONAL STRUCTURES AND PROPERTIES OF A TRANSFORMING AND A NONTRANSFORMING GLYCINE-12 MUTANT OF P21H-RAS | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Scheidig, A.J, Krengel, U, Pai, E.F, Kabsch, W, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1993-03-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras.
Biochemistry, 32, 1993
|
|
5P21
 
 | |
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q4S
 
 | |
8QOB
 
 | |
1JAH
 
 | | H-RAS P21 PROTEIN MUTANT G12P, COMPLEXED WITH GUANOSINE-5'-[BETA,GAMMA-METHYLENE] TRIPHOSPHATE AND MAGNESIUM | | Descriptor: | C-HA-RAS, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Schweins, T, Scheffzek, K, Assheuer, R, Wittinghofer, A. | | Deposit date: | 1996-12-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of the metal ion in the p21ras catalysed GTP-hydrolysis: Mn2+ versus Mg2+.
J.Mol.Biol., 266, 1997
|
|
1JAI
 
 | | H-RAS P21 PROTEIN MUTANT G12P, COMPLEXED WITH GUANOSINE-5'-[BETA,GAMMA-METHYLENE] TRIPHOSPHATE AND MANGANESE | | Descriptor: | C-HA-RAS, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Schweins, T, Scheffzek, K, Assheuer, R, Wittinghofer, A. | | Deposit date: | 1996-12-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of the metal ion in the p21ras catalysed GTP-hydrolysis: Mn2+ versus Mg2+.
J.Mol.Biol., 266, 1997
|
|
1AGP
 
 | | THREE-DIMENSIONAL STRUCTURES AND PROPERTIES OF A TRANSFORMING AND A NONTRANSFORMING GLY-12 MUTANT OF P21-H-RAS | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Franken, S.M, Scheidig, A.J, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1993-03-29 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras.
Biochemistry, 32, 1993
|
|
9GGP
 
 | | Alpha-1-antitrypsin in complex with the Fab fragment of an anti-polymer antibody | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1-antitrypsin, Fab fragment heavy chain of 2C1 monoclonal antibody, ... | | Authors: | Lowen, S.M, Laffranchi, M, Lomas, D.A, Irving, J.A. | | Deposit date: | 2024-08-13 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-resolution characterization of ex vivo AAT polymers by solution-state NMR spectroscopy.
Sci Adv, 11, 2025
|
|
2W3T
 
 | | Chloro complex of the Ni-Form of E.coli deformylase | | Descriptor: | CHLORIDE ION, ETHANOL, NICKEL (II) ION, ... | | Authors: | Ngo, Y.H.T, Palm, G.J, Hinrichs, W. | | Deposit date: | 2008-11-14 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure of the Ni(II) Complex of Escherichia Coli Peptide Deformylase and Suggestions on Deformylase Activities Depending on Different Metal(II) Centres.
J.Biol.Inorg.Chem., 15, 2010
|
|
