7B92
 
 | |
7B9C
 
 | |
7B91
 
 | |
6QUS
 
 | | HsCKK (human CAMSAP1) decorated 13pf taxol-GDP microtubule | | Descriptor: | Calmodulin-regulated spectrin-associated protein 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J.M, Luo, Y, Xiang, S, Yang, C, Jiang, K, Stangier, M, Vemu, A, Cook, A, Wang, S, Roll-Mecak, A, Steinmetz, M.O, Akhmanova, A, Baldus, M, Moores, C.A. | | Deposit date: | 2019-02-28 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural determinants of microtubule minus end preference in CAMSAP CKK domains.
Nat Commun, 10, 2019
|
|
7B0I
 
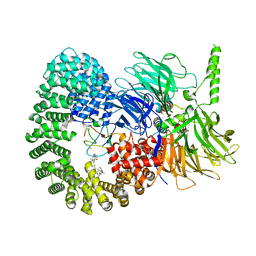 | | Structure of a minimal SF3B core in complex with spliceostatin A (form II) | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3,Splicing factor 3B subunit 3,Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2020-11-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
5ZYA
 
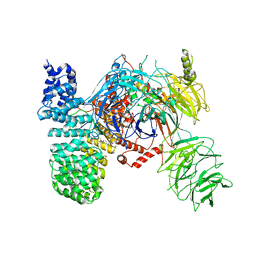 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
7KIA
 
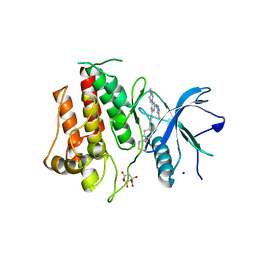 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 19 | | Descriptor: | 1-[4-(4-{4-(4-methylpiperazin-1-yl)-6-[(3-methyl-1H-pyrazol-5-yl)amino]pyrimidin-2-yl}phenyl)piperidin-1-yl]prop-2-en-1-one, CITRATE ANION, Fibroblast growth factor receptor 2, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
7KIE
 
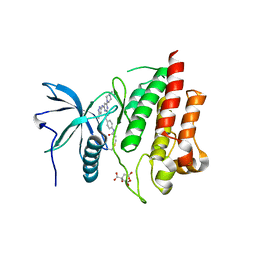 | | Crystal structure of FGFR2 kinase domain gatekeeper mutant V564F in complex with covalent compound 3 | | Descriptor: | CITRATE ANION, Fibroblast growth factor receptor 2, GLYCEROL, ... | | Authors: | Ke, J, Wibowo, A.S, Carter, J.J, Larsen, N.A. | | Deposit date: | 2020-10-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Discovery of Aminopyrazole Derivatives as Potent Inhibitors of Wild-Type and Gatekeeper Mutant FGFR2 and 3.
Acs Med.Chem.Lett., 12, 2021
|
|
6W2J
 
 | | CPS1 bound to allosteric inhibitor H3B-374 | | Descriptor: | (2-fluoranyl-4-methoxy-phenyl)-[(3~{R},5~{R})-4-(2-fluoranyl-4-methoxy-phenyl)carbonyl-3,5-dimethyl-piperazin-1-yl]methanone, 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2020-03-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of 2,6-Dimethylpiperazines as Allosteric Inhibitors of CPS1.
Acs Med.Chem.Lett., 11, 2020
|
|
7OPI
 
 | |
7ONB
 
 | | Structure of the U2 5' module of the A3'-SSA complex | | Descriptor: | MINX, PHD finger-like domain-containing protein 5A, RNU2, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
7OMF
 
 | | Structure of a minimal SF3B core in complex with sudemycin D6 (form I) | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
4CCU
 
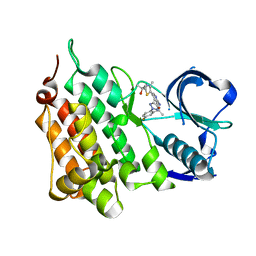 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy) pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol | | Descriptor: | 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propan-2-ol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4CCB
 
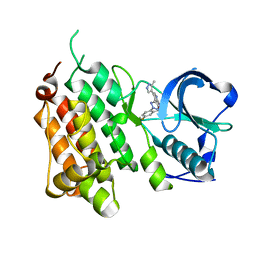 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with 3-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2-yl)phenyl)ethoxy)-5-(5-methyl-1H- pyrazol-4-yl)pyridin-2-amine | | Descriptor: | 3-[(1R)-1-[5-fluoranyl-2-(1,2,3-triazol-2-yl)phenyl]ethoxy]-5-(3-methyl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
4CD0
 
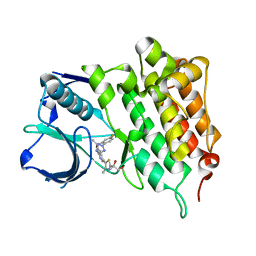 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with 2-(5-(6-amino-5-((R)-1-(5-fluoro-2-(2H-1,2,3-triazol-2- yl)phenyl)ethoxy)pyridin-3-yl)-4-methylthiazol-2-yl)propane-1,2-diol | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | The Design of Potent and Selective Inhibitors to Overcome Clinical Alk Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
8SOR
 
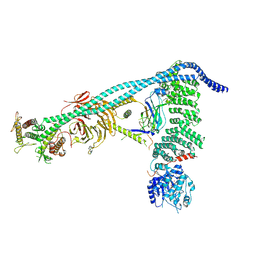 | | Structure of human PI3KC3-C1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beclin 1-associated autophagy-related key regulator, Beclin-1, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2023-04-29 | | Release date: | 2023-06-21 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
8SOI
 
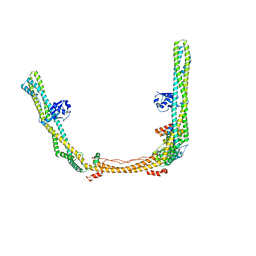 | | Structure of human ULK1 complex core (2:1:1 stoichiometry) | | Descriptor: | Autophagy-related protein 13, RB1-inducible coiled-coil protein 1, Serine/threonine-protein kinase ULK1 | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2023-04-28 | | Release date: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
4M0Y
 
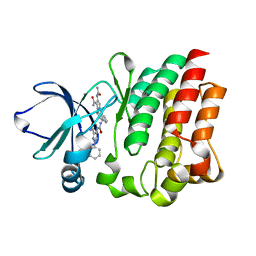 | |
4M13
 
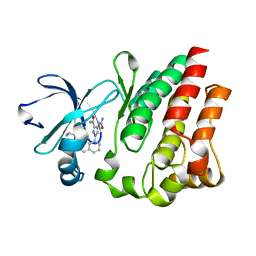 | |
4M0Z
 
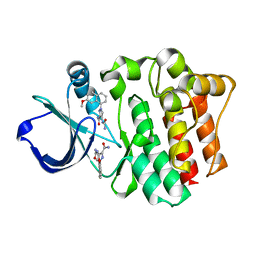 | |
4M12
 
 | |
8W1D
 
 | | CRYSTAL STRUCTURE OF DPS-LIKE PROTEIN PA4880 FROM PSEUDOMONAS AERUGINOSA (DIMERIC FORM) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION | | Authors: | Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8W1F
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer, Mg bound) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8W1E
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, SULFATE ION | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
4M14
 
 | |
