6Z5D
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004082 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 11-methyl-8,11,14,18,19,22-hexazatetracyclo[13.5.2.12,6.018,21]tricosa-1(21),2(23),3,5,15(22),16,19-heptaen-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2004082
To Be Published
|
|
4XWX
 
 | | Crystal structure of the PTB domain of SHC | | Descriptor: | 1,2-ETHANEDIOL, SHC-transforming protein 1, SODIUM ION | | Authors: | Chaikuad, A, Tallant, C, Krojer, T, Dixon-Clarke, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-01-29 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the PTB domain of SHC
To Be Published
|
|
3SOC
 
 | | Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with a quinazolin | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-2A, [4-({4-[(5-CYCLOPROPYL-1H-PYRAZOL-3-YL)AMINO]QUINAZOLIN-2-YL}IMINO)CYCLOHEXA-2,5-DIEN-1-YL]ACETONITRILE | | Authors: | Chaikuad, A, Williams, E, Mahajan, P, Cooper, C.D.O, Sanvitale, C, Vollmar, M, Muniz, J.R.C, Yue, W.W, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Activin receptor type-IIA (ACVR2A) kinase domain in complex with a quinazolin
To be Published
|
|
3MDY
 
 | | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Bone morphogenetic protein receptor type-1B, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Chaikuad, A, Sanvitale, C, Mahajan, P, Daga, N, Cooper, C, Krojer, T, Alfano, I, Knapp, S, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189
To be Published
|
|
4NR2
 
 | | Crystal structure of STK4 (MST1) SARAH domain | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 4 | | Authors: | Chaikuad, A, Krojer, T, Kopec, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of STK4 (MST1) SARAH domain
To be Published
|
|
5MUF
 
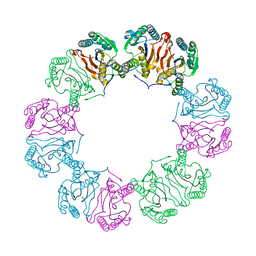 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in its enzymatically active dodecameric form induced by the presence of the N-terminal WDPNWD motif | | Descriptor: | PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
3MXO
 
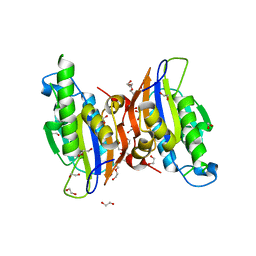 | | Crystal structure oh human phosphoglycerate mutase family member 5 (PGAM5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
7OPG
 
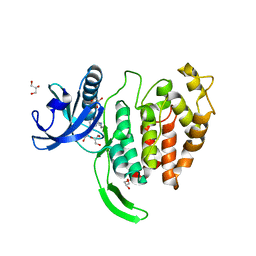 | | Crystal structure of CLK1 in complex with compound 2 (CC513) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[2-(propylamino)imidazo[2,1-b][1,3,4]thiadiazol-5-yl]phenol, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Routier, S, Bonnet, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of CLK1 in complex with compound 2 (CC513)
To Be Published
|
|
3U2U
 
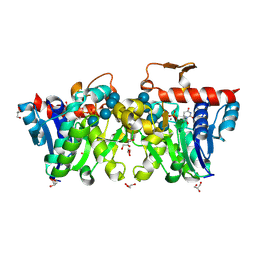 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltotetraose | | Descriptor: | GLYCEROL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U2W
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and glucose or a glucal species | | Descriptor: | 1,5-anhydro-D-arabino-hex-1-enitol, GLYCEROL, Glycogenin-1, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T7M
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a triclinic closed form | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T7O
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP-Glucose and glucose | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3T7N
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP, in a monoclinic closed form | | Descriptor: | Glycogenin-1, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U2T
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U2X
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and 1'-deoxyglucose | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-D-glucitol, Glycogenin-1, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and 1'-deoxyglucose
To be Published
|
|
3U2V
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltohexaose | | Descriptor: | GLYCEROL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3LWK
 
 | | Crystal structure of human Beta-crystallin A4 (CRYBA4) | | Descriptor: | Beta-crystallin A4, GLYCEROL, PHOSPHATE ION | | Authors: | Chaikuad, A, Shafqat, N, Krojer, T, Yue, W.W, Cocking, R, Vollmar, M, Muniz, J.R.C, Pike, A.C.W, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Beta-crystallin A4 (CRYBA4)
To be Published
|
|
3MAO
 
 | | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1) | | Descriptor: | FE (III) ION, MALONATE ION, Methionine-R-sulfoxide reductase B1, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Savitsky, P, Krojer, T, Ugochukwu, E, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1)
To be Published
|
|
7QS1
 
 | |
7QRZ
 
 | |
7QS2
 
 | |
7QRW
 
 | |
7QS4
 
 | |
7QS5
 
 | |
7QRY
 
 | |
