3SDP
 
 | |
5GCH
 
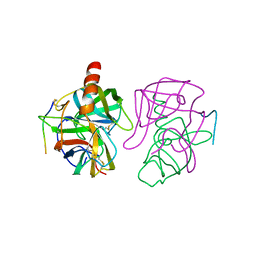 | |
3VGN
 
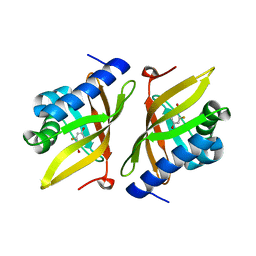 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 3-fluoro-4-nitrophenol | | Descriptor: | 3-fluoro-4-nitrophenol, Steroid Delta-isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P.A. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Quantitative dissection of hydrogen bond-mediated proton transfer in the ketosteroid isomerase active site
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3FZW
 
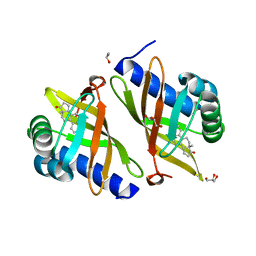 | | Crystal Structure of Ketosteroid Isomerase D40N-D103N from Pseudomonas putida (pKSI) with bound equilenin | | Descriptor: | EQUILENIN, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Ringe, D, Petsko, G.A. | | Deposit date: | 2009-01-26 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Hydrogen bond coupling in the ketosteroid isomerase active site.
Biochemistry, 48, 2009
|
|
2SFP
 
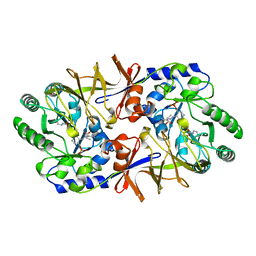 | | ALANINE RACEMASE WITH BOUND PROPIONATE INHIBITOR | | Descriptor: | PROPANOIC ACID, PROTEIN (ALANINE RACEMASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Morollo, A.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-02-16 | | Release date: | 1999-02-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Michaelis complex analogue: propionate binds in the substrate carboxylate site of alanine racemase.
Biochemistry, 38, 1999
|
|
6XNT
 
 | | Crystal structure of I91A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XNO
 
 | | Crystal structure of E99A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XNW
 
 | | Crystal structure of V39A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, NICKEL (II) ION | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XO1
 
 | | Crystal structure of N97A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
1BKH
 
 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BRM
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 1998-08-24 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis.
J.Mol.Biol., 289, 1999
|
|
1CM7
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1CNZ
 
 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | MANGANESE (II) ION, PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE), SULFATE ION | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-24 | | Release date: | 1999-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of Eschericia Coli and Salmonella Typhimurium 3-
Isopropylmalate Dehydrogenase and Comparison with Their Thermophilic
Counterpart from Thermus Thermophilus
J.Mol.Biol., 266, 1997
|
|
1TPC
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1CP6
 
 | | 1-BUTANEBORONIC ACID BINDING TO AEROMONAS PROTEOLYTICA AMINOPEPTIDASE | | Descriptor: | 1-BUTANE BORONIC ACID, PROTEIN (AMINOPEPTIDASE), ZINC ION | | Authors: | Depaola, C.C, Bennett, B, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development.
Biochemistry, 38, 1999
|
|
1YPI
 
 | |
6C2Z
 
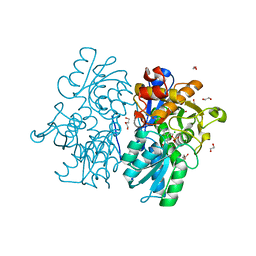 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-Aminoacrylate Intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
6C2Q
 
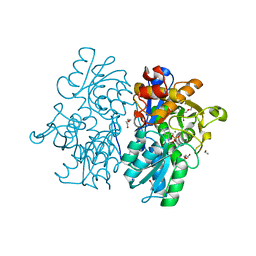 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-L-Serine Intermediate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
6C2H
 
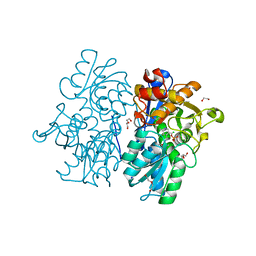 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the Catalytic Core | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
6C4P
 
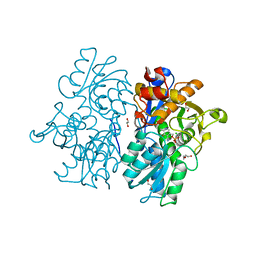 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PMP Complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Berkowitz, D.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
1MRA
 
 | |
3IAE
 
 | | Structure of benzaldehyde lyase A28S mutant with benzoylphosphonate | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzaldehyde lyase, CALCIUM ION | | Authors: | Brandt, G.S, Petsko, G.A, Ringe, D, McLeish, M.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site engineering of benzaldehyde lyase shows that a point mutation can confer both new reactivity and susceptibility to mechanism-based inhibition.
J.Am.Chem.Soc., 132, 2010
|
|
3IAF
 
 | | Structure of benzaldehyde lyase A28S mutant with monomethyl benzoylphosphonate | | Descriptor: | Benzaldehyde lyase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Brandt, G.S, Petsko, G.A, Ringe, D, McLeish, M.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Active-site engineering of benzaldehyde lyase shows that a point mutation can confer both new reactivity and susceptibility to mechanism-based inhibition.
J.Am.Chem.Soc., 132, 2010
|
|
3LZ2
 
 | | STRUCTURE DETERMINATION OF TURKEY EGG WHITE LYSOZYME USING LAUE DIFFRACTION | | Descriptor: | TURKEY EGG WHITE LYSOZYME | | Authors: | Howell, P.L, Almo, S.C, Parsons, M.R, Hajdu, J, Petsko, G.A. | | Deposit date: | 1991-09-13 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination of turkey egg-white lysozyme using Laue diffraction data.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
1DTN
 
 | |
