5UP0
 
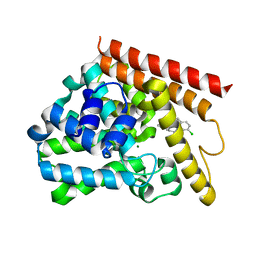 | | Crystal structure of human PDE1B catalytic domain in complex with inhibitor 3 (6-(4-chlorobenzyl)-8,9,10,11-tetrahydrobenzo[4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one) | | Descriptor: | 6-[(4-chlorophenyl)methyl]-8,9,10,11-tetrahydro[1]benzothieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, MAGNESIUM ION, ... | | Authors: | Cedervall, E.P, Allerston, C.K, Xu, R, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-01 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
8Y63
 
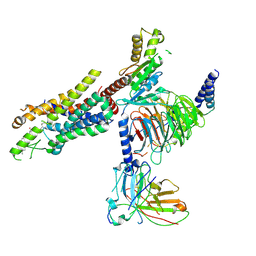 | | Cryo-EM structure of the C20:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Cer(d18:0/20:0), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 388, 2025
|
|
8Y62
 
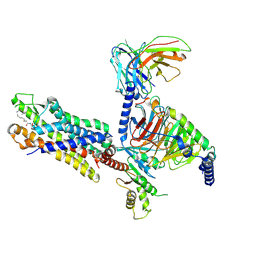 | | Cryo-EM structure of the C16:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 388, 2025
|
|
5X5L
 
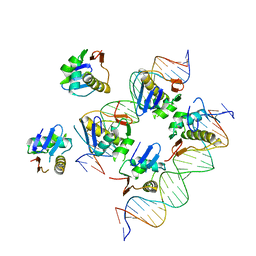 | | Crystal structure of response regulator AdeR DNA binding domain in complex with an intercistronic region | | Descriptor: | AdeR, DNA (5'-D(P*TP*AP*AP*AP*GP*TP*GP*TP*GP*GP*AP*GP*TP*AP*AP*GP*TP*GP*TP*GP*GP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*CP*CP*AP*CP*AP*CP*TP*TP*AP*CP*TP*CP*CP*AP*CP*AP*CP*TP*TP*TP*A)-3') | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5X5J
 
 | | Crystal structure of response regulator AdeR receiver domain | | Descriptor: | AdeR | | Authors: | Wen, Y. | | Deposit date: | 2017-02-16 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
5UWF
 
 | | Crystal structure of human PDE10A in complex with inhibitor 16d | | Descriptor: | 9-[(1S)-2,2-difluorocyclopropane-1-carbonyl]-6-[(4-methoxyphenyl)methyl]-8,9,10,11-tetrahydropyrido[4',3':4,5]thieno[3,2-e][1,2,4]triazolo[1,5-c]pyrimidin-5(6H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Xu, R, Cedervall, E.P, Sridhar, V, Barker, R, Aertgeerts, K. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Selective Phosphodiesterase 1 Inhibitors with Memory Enhancing Properties.
J. Med. Chem., 60, 2017
|
|
8HPM
 
 | | LpqY-SugABC in state 2 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPN
 
 | | LpqY-SugABC in state 3 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPS
 
 | | LpqY-SugABC in state 5 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPL
 
 | | LpqY-SugABC in state 1 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPR
 
 | | LpqY-SugABC in state 4 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
7EZG
 
 | | The structure of the human METTL6 enzyme in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Xie, W, Chen, R, Zhou, J, Liu, L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human METTL6, the m 3 C methyltransferase.
Commun Biol, 4, 2021
|
|
5XJP
 
 | | Crystal structure of response regulator AdeR receiver domain with Mg | | Descriptor: | AdeR, MAGNESIUM ION | | Authors: | Wen, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region.
Nucleic Acids Res., 45, 2017
|
|
6A4C
 
 | | Solution structure of MXAN_0049 | | Descriptor: | Uncharacterized protein MXAN_0049 | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MXAN_0049
To Be Published
|
|
7WQH
 
 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
8HQG
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQI
 
 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQH
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with inhibitor YH-53 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8HQF
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Non-structural protein 7 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2024-01-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
7VVP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
6BQS
 
 | | HusA haemophore from Porphyromonas gingivalis | | Descriptor: | hypothetical protein PG_2227 | | Authors: | Gell, D.A, Kwan, A.H, Horne, J, Hugrass, B.M, Collins, D.A.T. | | Deposit date: | 2017-11-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural properties of a haemophore facilitate targeted elimination of the pathogen Porphyromonas gingivalis.
Nat Commun, 9, 2018
|
|
8X8K
 
 | | Crystal structure of STBD1 CBM20 domain in complex with maltotetraose | | Descriptor: | GLYCEROL, Starch-binding domain-containing protein 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WUR
 
 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhao, Z.Y, Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2023-10-21 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
7VFB
 
 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFA
 
 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
