8KEJ
 
 | |
8KEP
 
 | |
8KDT
 
 | |
8KEK
 
 | |
8KDR
 
 | |
8KEQ
 
 | |
8KDS
 
 | |
8KEO
 
 | |
8KER
 
 | |
8KEH
 
 | |
8H08
 
 | |
5KR1
 
 | | Protease PR5-DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease PR5-DRV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5INW
 
 | | Structure of reaction loop cleaved lamprey angiotensinogen | | Descriptor: | C-terminal peptide of Putative angiotensinogen, Putative angiotensinogen, SULFATE ION | | Authors: | Wei, H, Zhou, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heparin Binds Lamprey Angiotensinogen and Promotes Thrombin Inhibition through a Template Mechanism
J.Biol.Chem., 291, 2016
|
|
8H07
 
 | |
5KQX
 
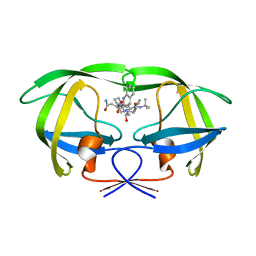 | | Protease E35D-SQV | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease E35D-SQV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KR0
 
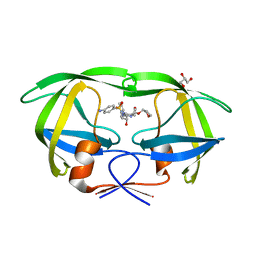 | | Protease E35D-APV | | Descriptor: | GLYCEROL, Protease E35D-APV, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5V0N
 
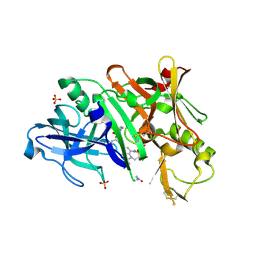 | | BACE1 in complex with inhibitor 5g | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{(1S,2S)-1-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-hydroxy-3-phenylpropan-2-yl}-7-ethyl-1,3,3-trimethyl-2,2-dioxo-1,2,3,4-tetrahydro-2lambda~6~-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide, ... | | Authors: | Mesecar, A, Ghosh, A, Yen, Y.-C. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Design, synthesis, and X-ray structural studies of BACE-1 inhibitors containing substituted 2-oxopiperazines as P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5KR2
 
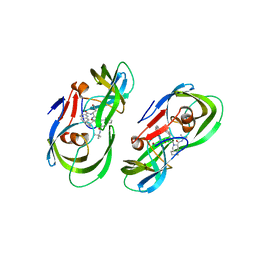 | | Protease PR5-SQV | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease PR5-SQV | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
5KZI
 
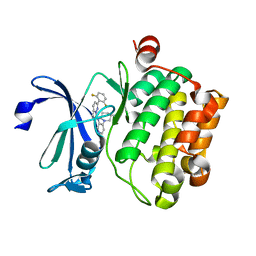 | | Crystal structure of human Pim-1 kinase in complex with an imidazopyridazine inhibitor. | | Descriptor: | Serine/threonine-protein kinase pim-1, ~{N}-[4-[(3~{S})-3-azanylpiperidin-1-yl]pyridin-3-yl]-2-[2,6-bis(fluoranyl)phenyl]imidazo[1,5-b]pyridazin-7-amine | | Authors: | Mohr, C. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of imidazopyridazines as potent Pim-1/2 kinase inhibitors.
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5VMR
 
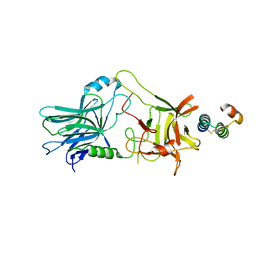 | |
5VSW
 
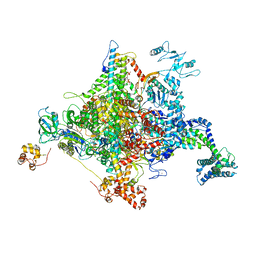 | |
5VID
 
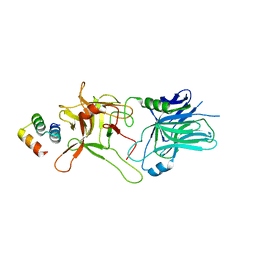 | |
5VLI
 
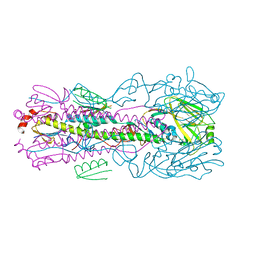 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
4FA6
 
 | |
5GY3
 
 | |
