1H65
 
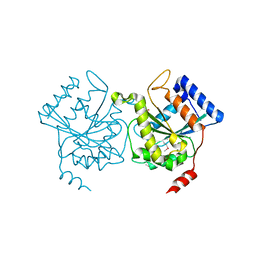 | | Crystal structure of pea Toc34 - a novel GTPase of the chloroplast protein translocon | | Descriptor: | CHLOROPLAST OUTER ENVELOPE PROTEIN OEP34, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Sun, Y.J, Forouhar, F, Li, H.M, Tu, S.L, Kao, S, Shr, H.L, Chou, C.C, Hsiao, C.D. | | Deposit date: | 2001-06-06 | | Release date: | 2002-01-29 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Pea Toc34 - a Novel Gtpase of the Chloroplast Protein Translocon
Nat.Struct.Biol., 9, 2002
|
|
1HEV
 
 | |
7VNE
 
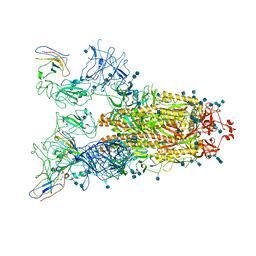 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113.1 (UUU-state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VND
 
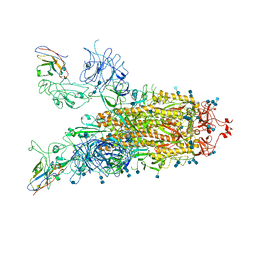 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UUD-state, state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with a human single domain antibody n3113 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, n3113 | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
7VNC
 
 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with a human single domain antibody n3113 (UDD-state, state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, Z, Wang, Y, Kong, Y, Jin, Y, Wu, Y, Ying, T. | | Deposit date: | 2021-10-10 | | Release date: | 2021-11-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A non-ACE2 competing human single-domain antibody confers broad neutralization against SARS-CoV-2 and circulating variants.
Signal Transduct Target Ther, 6, 2021
|
|
4N2S
 
 | | Crystal Structure of THA8 in complex with Zm1a-6 RNA | | Descriptor: | THA8 RNA binding protein, Zm1a-6 RNA | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4N2Q
 
 | | Crystal structure of THA8 in complex with Zm4 RNA | | Descriptor: | RNA (5'-R(*AP*AP*GP*AP*AP*GP*AP*AP*AP*UP*UP*GP*G)-3'), THA8 RNA binding protein | | Authors: | Ke, J, Chen, R.Z, Ban, T, Zhou, X.E, Gu, X, Brunzelle, J.S, Zhu, J.K, Melcher, K, Xu, H.E. | | Deposit date: | 2013-10-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA recognition by a dimeric PPR-protein complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3F1Z
 
 | |
3C7L
 
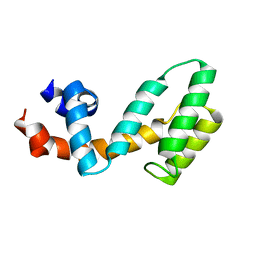 | | Molecular architecture of Galphao and the structural basis for RGS16-mediated deactivation | | Descriptor: | Regulator of G-protein signaling 16 | | Authors: | Slep, K.C, Kercher, M.A, Wieland, T, Chen, C, Simon, M.I, Sigler, P.B. | | Deposit date: | 2008-02-07 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular architecture of G{alpha}o and the structural basis for RGS16-mediated deactivation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6W9H
 
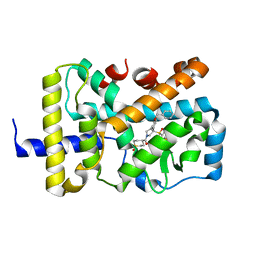 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | 4-{(3R)-3-[4-(benzyloxy)phenyl]-3-[(4-fluorophenyl)sulfonyl]pyrrolidine-1-carbonyl}-1lambda~6~-thiane-1,1-dione, Nuclear receptor ROR-gamma, Steroid receptor coactivator 1 fusion | | Authors: | Sack, J.S. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6W9I
 
 | |
6XAE
 
 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | Nuclear receptor ROR-gamma, trans-4-{(3aR,9bR)-7-[(2-chloro-6-fluorophenyl)methoxy]-9b-[(4-fluorophenyl)sulfonyl]-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl}cyclohexane-1-carboxylic acid | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4CSO
 
 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
4OA7
 
 | | Crystal structure of Tankyrase1 in complex with IWR1 | | Descriptor: | 4-[(3aR,4R,7S,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Zhang, X, He, H. | | Deposit date: | 2014-01-03 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
2OBK
 
 | | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens. Northeast Structural Genomics Consortium target PlR6. | | Descriptor: | SelT/selW/selH selenoprotein domain | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-19 | | Release date: | 2007-01-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-Ray structure of the putative Se binding protein from Pseudomonas fluorescens
To be Published
|
|
2O1M
 
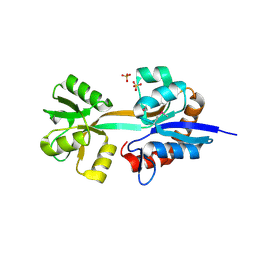 | | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572 | | Descriptor: | Probable amino-acid ABC transporter extracellular-binding protein ytmK, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the probable amino-acid ABC transporter extracellular-binding protein ytmK from Bacillus subtilis. Northeast Structural Genomics Consortium target SR572.
To be Published
|
|
6M1K
 
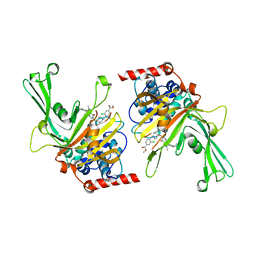 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
5WJ8
 
 | | Crystal Structure of Human Cadherin-23 EC13-14 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cadherin-23, ... | | Authors: | Velez-Cortes, F, Conghui, C, De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-07-21 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
4QTO
 
 | | 1.65 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) from Staphylococcus aureus with BME-modified Cys289 and PEG molecule in active site | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine aldehyde dehydrogenase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-08 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4I1L
 
 | | Structural and Biological Features of FOXP3 Dimerization Relevant to Regulatory T Cell Function | | Descriptor: | ACETATE ION, Forkhead box protein P3, MAGNESIUM ION, ... | | Authors: | Song, X.M, Greene, M.I, Zhou, Z.C. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function.
Cell Rep, 1, 2012
|
|
2M7T
 
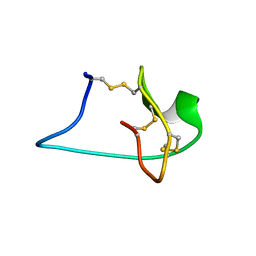 | | Solution NMR Structure of Engineered Cystine Knot Protein 2.5D | | Descriptor: | Cystine Knot Protein 2.5D | | Authors: | Cochran, F.V, Das, R. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
4S05
 
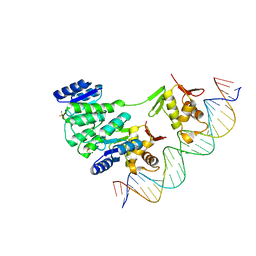 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (26-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
4S04
 
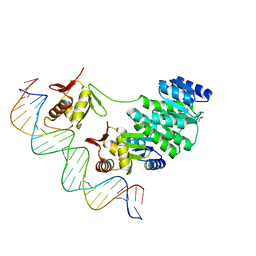 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (25-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
4LGI
 
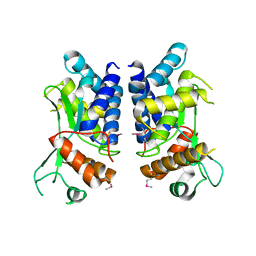 | | N-terminal truncated NleC structure | | Descriptor: | Uncharacterized protein | | Authors: | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-01-15 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
