8AKN
 
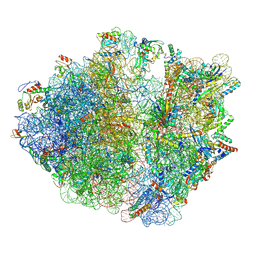 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-07-30 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
8ANA
 
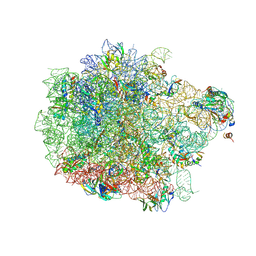 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the 50S ribosomal subunit | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
8AM9
 
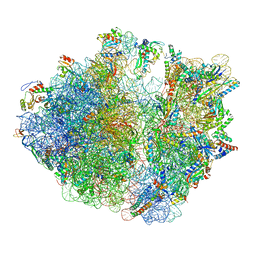 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the elongating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
5A06
 
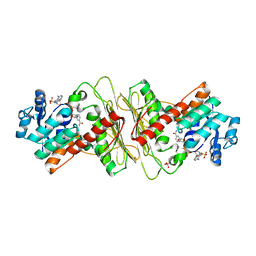 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with sorbitol | | Descriptor: | ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
5A03
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with xylose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
5A05
 
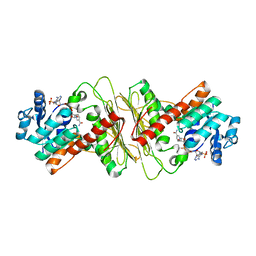 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with maltotriose | | Descriptor: | ALDOSE-ALDOSE OXIDOREDUCTASE, MALONATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
5A04
 
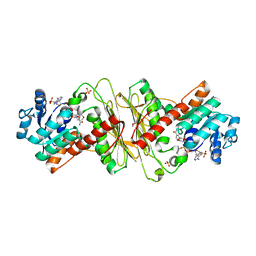 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glucose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ALDOSE-ALDOSE OXIDOREDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
6G5J
 
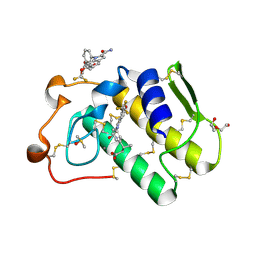 | | Secreted phospholipase A2 type X in complex with ligand | | Descriptor: | (3~{R})-3-[3-[2-aminocarbonyl-6-(trifluoromethyloxy)indol-1-yl]phenyl]butanoic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Selective sPLA2-X Inhibitor (-)-2-{2-[Carbamoyl-6-(trifluoromethoxy)-1H-indol-1-yl]pyridine-2-yl}propanoic Acid.
ACS Med Chem Lett, 9, 2018
|
|
8ACK
 
 | |
8ACR
 
 | |
8ACG
 
 | |
8AC7
 
 | |
8AC9
 
 | |
5A02
 
 | | Crystal structure of aldose-aldose oxidoreductase from Caulobacter crescentus complexed with glycerol | | Descriptor: | ALDOSE-ALDOSE OXIDOREDUCTASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Taberman, H, Rouvinen, J, Parkkinen, T. | | Deposit date: | 2015-04-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Caulobacter Crescentus Aldose-Aldose Oxidoreductase.
Biochem.J., 472, 2015
|
|
8SU8
 
 | |
8T7V
 
 | |
8T09
 
 | |
3JB4
 
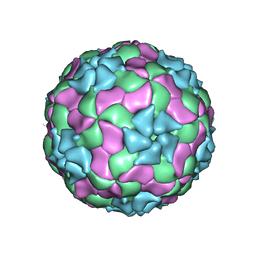 | | Structure of Ljungan virus: insight into picornavirus packaging | | Descriptor: | VP0, VP1, VP3 | | Authors: | Zhu, L, Wang, X.X, Ren, J.S, Porta, C, Wenham, H, Ekstrom, J.-O, Panjwani, A, Knowles, N.J, Kotecha, A, Siebert, A, Lindberg, M, Fry, E.E, Rao, Z.H, Tuthill, T.J, Stuart, D.I. | | Deposit date: | 2015-07-21 | | Release date: | 2015-10-21 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Ljungan virus provides insight into genome packaging of this picornavirus.
Nat Commun, 6, 2015
|
|
1U6Q
 
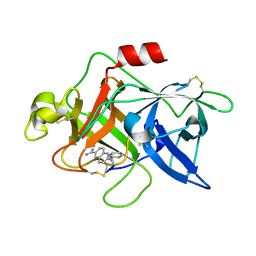 | | Substituted 2-Naphthamadine inhibitors of Urokinase | | Descriptor: | TRANS-6-(2-PHENYLCYCLOPROPYL)-NAPHTHALENE-2-CARBOXAMIDINE, Urokinase-type plasminogen activator | | Authors: | Bruncko, M, McClellan, W, Wendt, M.D, Sauer, D.R, Geyer, A, Dalton, C.R, Kaminski, M.K, Nienaber, V.L, Rockway, T.R, Giranda, V.L. | | Deposit date: | 2004-07-30 | | Release date: | 2004-10-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Naphthamidine urokinase plasminogen activator inhibitors with improved pharmacokinetic properties.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3G7K
 
 | |
2J4R
 
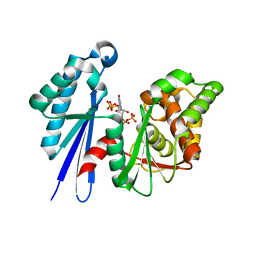 | | Structural Study of the Aquifex aeolicus PPX-GPPA enzyme | | Descriptor: | EXOPOLYPHOSPHATASE, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Kristensen, O. | | Deposit date: | 2006-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the Ppx/Gppa Phosphatase from Aquifex Aeolicus in Complex with the Alarmone Ppgpp
J.Mol.Biol., 375, 2008
|
|
2CFI
 
 | | The hydrolase domain of human 10-FTHFD in complex with 6- formyltetrahydropterin | | Descriptor: | 10-FORMYLTETRAHYDROFOLATE DEHYDROGENASE, 6-FORMYLTETRAHYDROPTERIN, SULFATE ION | | Authors: | Kursula, P, Stenmark, P, Arrowsmith, C, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Ogg, D.J, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Weigelt, J. | | Deposit date: | 2006-02-21 | | Release date: | 2006-03-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the Hydrolase Domain of Human 10-Formyltetrahydrofolate Dehydrogenase and its Complex with a Substrate Analogue.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BW0
 
 | | Crystal Structure of the hydrolase domain of Human 10-Formyltetrahydrofolate 2 dehydrogenase | | Descriptor: | 10-FORMYLTETRAHYDROFOLATE DEHYDROGENASE, SULFATE ION | | Authors: | Ogg, D.J, Stenmark, P, Arrowsmith, C, Edwards, A, Ehn, M, Graslund, S, Hammarstrom, M, Hallberg, M, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Persson, C, Sagemark, J, Schuler, H, Sundstrom, M, Thorsell, A, Dobritzsch, D, Weigelt, J. | | Deposit date: | 2005-07-07 | | Release date: | 2005-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the Hydrolase Domain of Human 10-Formyltetrahydrofolate Dehydrogenase and its Complex with a Substrate Analogue.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7AMF
 
 | |
7AMB
 
 | |
