3EEG
 
 | |
3E9M
 
 | |
4DUP
 
 | | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42 | | Descriptor: | quinone oxidoreductase | | Authors: | Kumaran, D, Rice, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-22 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of a quinone oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
3EUW
 
 | |
3F5D
 
 | |
3FD8
 
 | |
3FII
 
 | |
3CK5
 
 | |
3CO8
 
 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of alanine racemase from Oenococcus oeni with bound pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3CYG
 
 | |
3D8K
 
 | |
3DEB
 
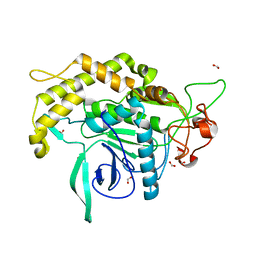 | |
3DLI
 
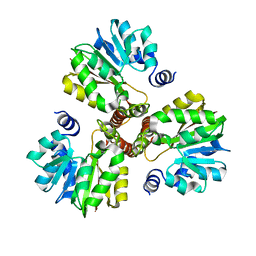 | |
3EWN
 
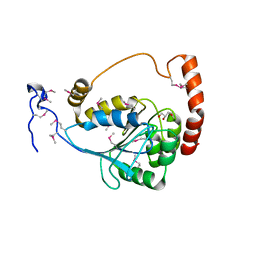 | |
3FJY
 
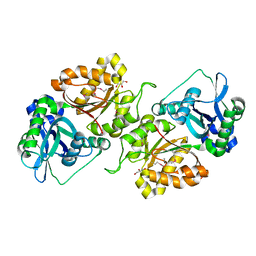 | |
3FIJ
 
 | |
3CMN
 
 | |
3D3X
 
 | |
3CVG
 
 | |
3D3A
 
 | |
3B89
 
 | |
3D5L
 
 | |
4E21
 
 | | The crystal structure of 6-phosphogluconate dehydrogenase from Geobacter metallireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 6-phosphogluconate dehydrogenase (Decarboxylating) | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-07 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | The crystal structure of 6-phosphogluconate dehydrogenase from Geobacter metallireducens
To be Published
|
|
3DMY
 
 | |
3DH0
 
 | |
