6JAK
 
 | | OtsA apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Wang, D, Liu, J. | | Deposit date: | 2019-01-24 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
7E2W
 
 | | Crystal structure of isocitrate dehydrogenase from Ostreococcus tauri in complex with isocitrate and magnesium(II) | | Descriptor: | CITRATE ANION, GLYCEROL, ISOCITRIC ACID, ... | | Authors: | Zhu, G.P, Tang, W.G, Wang, P. | | Deposit date: | 2021-02-07 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of NAD + -linked isocitrate dehydrogenase from the green alga Ostreococcus tauri and its evolutionary relationship with eukaryotic NADP + -linked homologs.
Arch.Biochem.Biophys., 708, 2021
|
|
6JBI
 
 | | Structure of Tps1 apo structure | | Descriptor: | Trehalose-6-phosphate synthase | | Authors: | Wang, S, Zhao, Y, Yi, L, Wang, D, Liu, J. | | Deposit date: | 2019-01-25 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Magnaporthe oryzae trehalose-6-phosphate synthase (MoTps1) suggest a model for catalytic process of Tps1.
Biochem.J., 476, 2019
|
|
8K6C
 
 | |
8K68
 
 | |
8K6A
 
 | |
8K6D
 
 | |
8K67
 
 | |
8K6B
 
 | |
6K9P
 
 | | Structure of Deubiquitinase | | Descriptor: | Ubiquitin, Ubiquitin thioesterase | | Authors: | Lu, L.N, Liu, L, Wang, F. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains
Nat Commun, 13, 2022
|
|
4UB0
 
 | |
8GBN
 
 | | Structure of Apo Human SIRT5 P114T Mutant | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8GBL
 
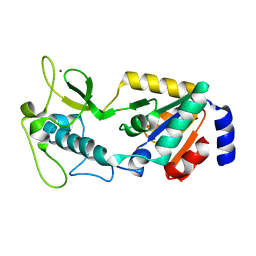 | | Structure of Apo Human SIRT5 | | Descriptor: | NAD-dependent protein deacylase sirtuin-5, mitochondrial, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
3R4N
 
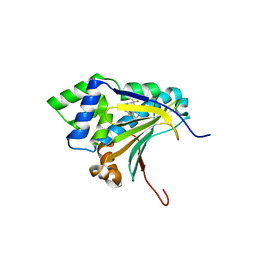 | | Optimization of Potent, Selective, and Orally Bioavailable Pyrrolodinopyrimidine-containing Inhibitors of Heat Shock Protein 90. Identification of Development Candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 4-[2-chloro-6-(4,4,4-trifluorobutoxy)phenyl]-6-methylpyrimidin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
7E56
 
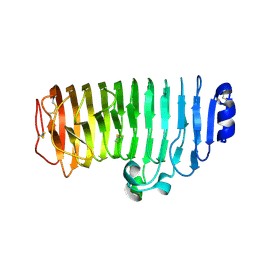 | |
7E4I
 
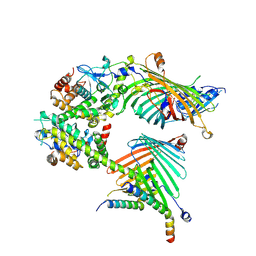 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40/Tom5/Tom6 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, Mitochondrial import receptor subunit TOM6, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7E4H
 
 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
6TF1
 
 | |
6TFF
 
 | |
6JGX
 
 | | Crystal structure of the transcriptional regulator CadR from P. putida in complex with Cadmium(II) and DNA | | Descriptor: | CADMIUM ION, CadR, DNA (5'-D(*CP*AP*CP*CP*CP*TP*AP*TP*AP*GP*TP*GP*GP*CP*TP*AP*CP*AP*GP*GP*GP*T)-3'), ... | | Authors: | Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Selective cadmium regulation mediated by a cooperative binding mechanism in CadR.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6TF0
 
 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TF3
 
 | |
6TFE
 
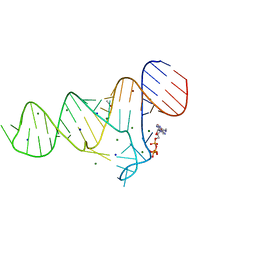 | |
6TNT
 
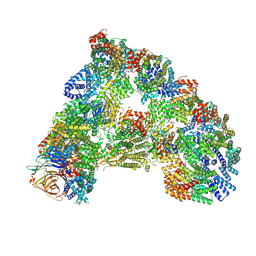 | | SUMOylated apoAPC/C with repositioned APC2 WHB domain | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Barford, D, Yatskevich, S. | | Deposit date: | 2019-12-10 | | Release date: | 2021-01-13 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular mechanisms of APC/C release from spindle assembly checkpoint inhibition by APC/C SUMOylation.
Cell Rep, 34, 2021
|
|
6JGW
 
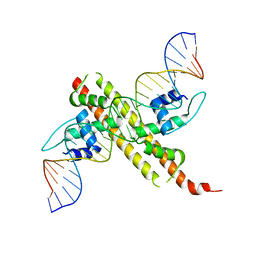 | |
