6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
7DMN
 
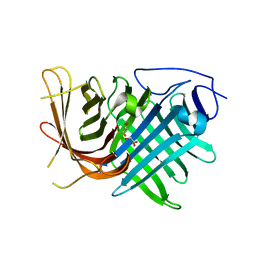 | | Crystal structure of two pericyclases catalyzing [4+2] cycloaddition | | Descriptor: | Diels-Alderase fsa2, GLYCEROL | | Authors: | Chi, C.B, Wang, Z.D, Liu, T, Zhang, Z.Y, Ma, M. | | Deposit date: | 2020-12-04 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Fsa2 and Phm7 Catalyzing [4 + 2] Cycloaddition Reactions with Reverse Stereoselectivities in Equisetin and Phomasetin Biosynthesis.
Acs Omega, 6, 2021
|
|
7DMO
 
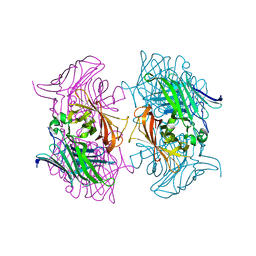 | |
7WE6
 
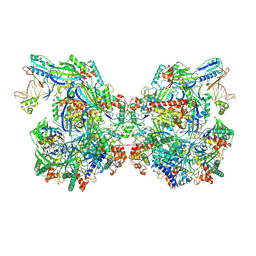 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
6LZG
 
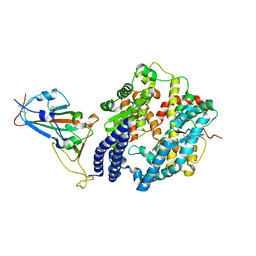 | | Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Wang, Q.H, Song, H, Qi, J.X. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2.
Cell, 181, 2020
|
|
7X29
 
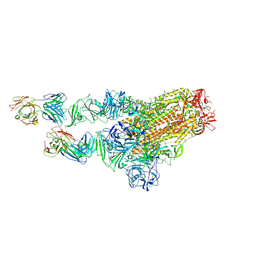 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X2A
 
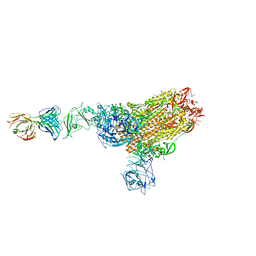 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X28
 
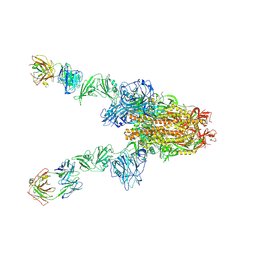 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X52
 
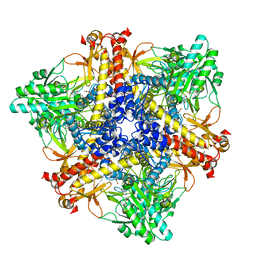 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP complex | | Descriptor: | ACETATE ION, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X4Y
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GABA complex | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X51
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GUA complex | | Descriptor: | GLUTARIC ACID, Glutamate decarboxylase, MALONATE ION, ... | | Authors: | Liu, S, Du, G, Wang, L, Wen, B, Xin, F. | | Deposit date: | 2022-03-03 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X4L
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | Deposit date: | 2022-03-02 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
5KW1
 
 | | Crystal Structure of the Two Tandem RRM Domains of PUF60 Bound to a Modified AdML Pre-mRNA 3' Splice Site Analogue | | Descriptor: | CHLORIDE ION, DNA/RNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KVY
 
 | | CRYSTAL STRUCTURE OF THE TWO TANDEM RRM DOMAINS OF PUF60 BOUND TO A PORTION OF AN ADML PRE-MRNA 3' SPLICE SITE ANALOG | | Descriptor: | CHLORIDE ION, DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Hsiao, H.-H, Crichlow, G.V, Albright, R.A, Murphy, J.W, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5KW6
 
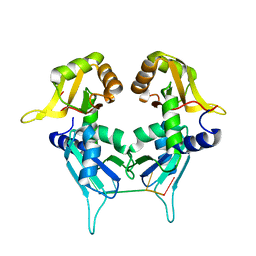 | | Two Tandem RRM Domains of PUF60 Bound to an AdML Pre-mRNA 3' Splice Site Analogue with a Modified Binding-Site Nucleic Acid Base | | Descriptor: | DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
7CJM
 
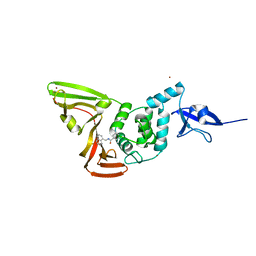 | | SARS CoV-2 PLpro in complex with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Fu, Z, Huang, H. | | Deposit date: | 2020-07-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
7DSY
 
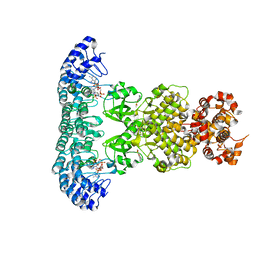 | | Crystal Structure of RNase L in complex with KM05073 | | Descriptor: | 1-chloranyl-3-methylsulfinyl-6,7-dihydro-5H-2-benzothiophen-4-one, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-03 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7DWO
 
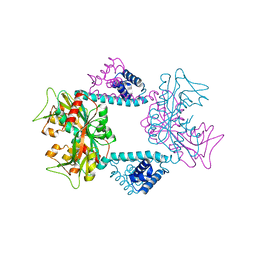 | |
7DWN
 
 | | Crystal structure of Vibrio fischeri DarR in complex with DNA reveals the transcriptional activation mechanism of LTTR family members | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Predicted DNA-binding transcriptional regulator | | Authors: | Wang, W.W, Wu, H, He, J.H, Yu, F. | | Deposit date: | 2021-01-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure details of Vibrio fischeri DarR and mutant DarR-M202I from LTTR family reveals their activation mechanism.
Int.J.Biol.Macromol., 183, 2021
|
|
7DTS
 
 | | Crystal structure of RNase L in complex with AC40357 | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7ELW
 
 | | Crystal structure of RNase L in complex with Myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
8IJZ
 
 | | anti-VEGF mutant | | Descriptor: | anti-VEGF nanobody | | Authors: | Qian, F, Zhu, S.Q. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
8IJS
 
 | | anti-VEGF nanobody mutant | | Descriptor: | ZINC ION, anti-VEGF nanobody | | Authors: | Qian, F, Zhu, S.Q. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
8IIU
 
 | | anti-VEGF nanobody | | Descriptor: | SULFATE ION, anti-VEGF nanobody | | Authors: | Qian, F, Zhu, S.Q. | | Deposit date: | 2023-02-24 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
