5KX8
 
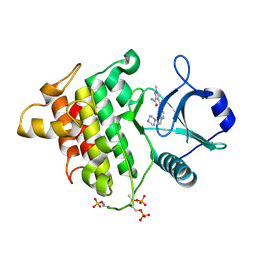 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(3-aminocarbonyl-1-methyl-pyrazol-4-yl)-5-piperazin-1-yl-pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.671 Å) | | Cite: | Efforts towards the optimization of a bi-aryl class of potent IRAK4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5KX7
 
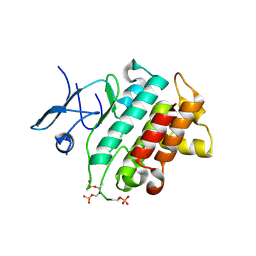 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N}-(3-aminocarbonyl-1-methyl-pyrazol-4-yl)-6-(1-methylpyrazol-4-yl)pyridine-2-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Efforts towards the optimization of a bi-aryl class of potent IRAK4 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5L9F
 
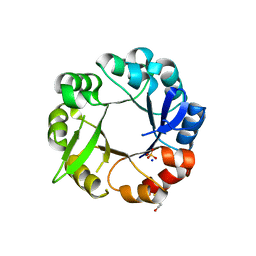 | | S. enterica HisA mutant - D10G, G11D, dup13-15, G44E, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SODIUM ION, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Selmer, M. | | Deposit date: | 2016-06-10 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LEV
 
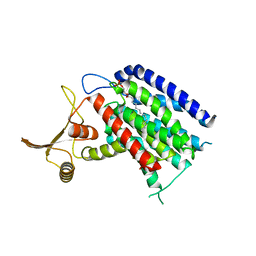 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) | | Descriptor: | UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, UNKNOWN LIGAND | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5MFA
 
 | | Crystal structure of human promyeloperoxidase (proMPO) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grishkovskaya, I, Furtmueller, P.G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human promyeloperoxidase (proMPO) and the role of the propeptide in processing and maturation.
J. Biol. Chem., 292, 2017
|
|
5O5E
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with tunicamycin | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
5L6U
 
 | | S. ENTERICA HISA MUTANT - D10G, DUP13-15, Q24L, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Selmer, M. | | Deposit date: | 2016-05-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NR8
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7a | | Descriptor: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-methyl-piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Podjarny, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
7N2O
 
 | | AS4.2-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2Q
 
 | | AS4.3-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2S
 
 | | AS3.1-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2R
 
 | | AS4.3-PRPF3-HLA*B27 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2N
 
 | | TCR-antigen complex AS4.2-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2P
 
 | | AS4.3-RNASEH2b-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
5G5J
 
 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
5GXG
 
 | | High-resolution crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kikui, Y, Hiruma, Y, Ubbink, M, Nojiri, M. | | Deposit date: | 2016-09-17 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of productive and futile encounters in an electron transfer protein complex
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6MBI
 
 | |
5G4W
 
 | | S. enterica HisA mutant D7N, D10G, Dup13-15 (VVR) with substrate ProFAR | | Descriptor: | GLYCEROL, HISA, [(2R,3S,4R,5R)-5-[4-aminocarbonyl-5-[(E)-[[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]amino]methylideneamino]imidazol-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G5I
 
 | | S. enterica HisA mutant D10G | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHOSPHATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-25 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1QJU
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP61209 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2N-METHYL-2H-TETRAZOL-5-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Minor, I, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-05 | | Release date: | 1999-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QJY
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND VP65099 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[2-METHYL-4-ISOXAZOLYL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QJX
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND WIN68934 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[4-METHYL-2H-TETRAZOL-2-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6NPZ
 
 | | Crystal structure of Akt1 (aa 123-480) kinase with a bisubstrate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | Authors: | Chu, N, Cole, P.A, Gabelli, S.B. | | Deposit date: | 2019-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
6NXJ
 
 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6NSI
 
 | | Crystal structure of Fe(III)-bound YtgA from Chlamydia trachomatis | | Descriptor: | CALCIUM ION, FE (III) ION, Manganese-binding protein, ... | | Authors: | Luo, Z, Campbell, R, Begg, S.L, Kobe, B, McDevitt, C.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.00006342 Å) | | Cite: | Structure and Metal Binding Properties of Chlamydia trachomatis YtgA.
J.Bacteriol., 202, 2019
|
|
