5UWY
 
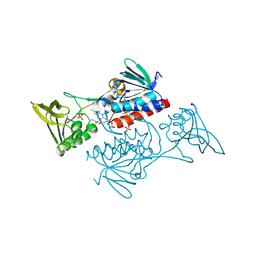 | | The crystal structure of thioredoxin reductase from Streptococcus pyogenes MGAS5005 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, Thioredoxin reductase | | Authors: | Tan, K, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-21 | | Release date: | 2017-03-15 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
5V0I
 
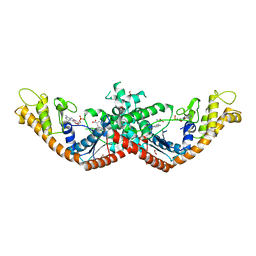 | | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, TRYPTOPHAN, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Grimshaw, S.G, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tryptophanyl-tRNA Synthetase from Escherichia coli Complexed with AMP and Tryptophan
To Be Published
|
|
5VSV
 
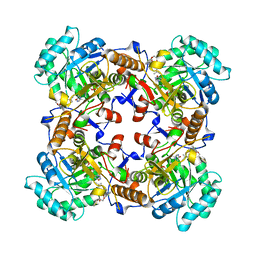 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P225 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, {2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenoxy}acetic acid | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P225
To Be Published
|
|
5VDN
 
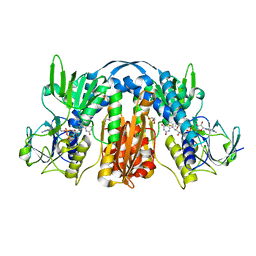 | | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Glutathione oxidoreductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD.
To Be Published
|
|
6E5Y
 
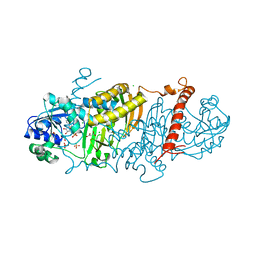 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
5WI5
 
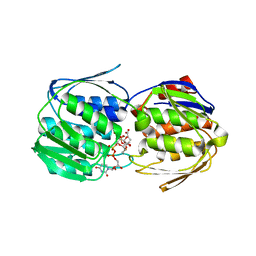 | | 2.0 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MAGNESIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-07-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Streptococcus pneumoniae in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
5UZE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P182
To Be Published
|
|
5UQH
 
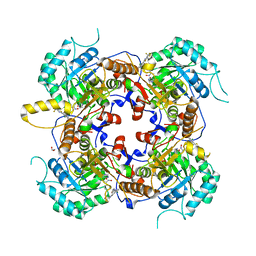 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p182 | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
5VH6
 
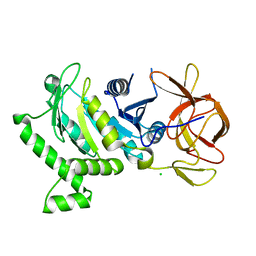 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
5DUL
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
5DLL
 
 | | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Aminopeptidase N, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Borek, D, Raczynska, J, Dubrovska, I, Grimshaw, S, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
5EAV
 
 | | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unliganded structure of the ornithine aminotransferase from Toxoplasma gondii
To Be Published
|
|
5E5I
 
 | | Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator | | Descriptor: | 4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-4-enoic acid, 6-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]-4-oxidanylidene-hexanoic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii in complex with inactivator.
To Be Published
|
|
5E3K
 
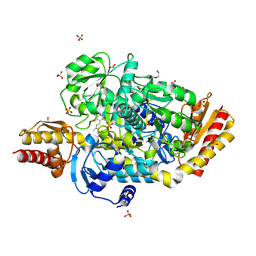 | | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid | | Descriptor: | 4-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-4-enoic acid, Aminotransferase, CARBONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-02 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with (S)-4-amino-5-fluoropentanoic acid
To Be Published
|
|
5DN8
 
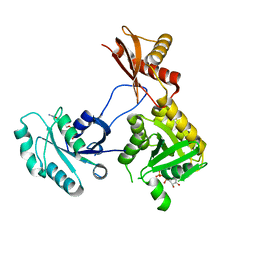 | | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP. | | Descriptor: | GTPase Der, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Minasov, G, Shuvalova, L, Han, A, Kim, H.-Y, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP.
To Be Published
|
|
5DZS
 
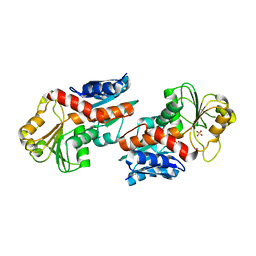 | | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile. | | Descriptor: | SULFATE ION, Shikimate dehydrogenase (NADP(+)) | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.
To Be Published
|
|
5E31
 
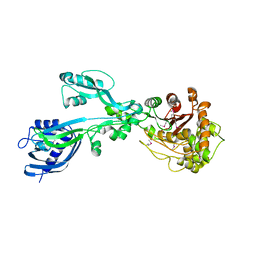 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5HJ1
 
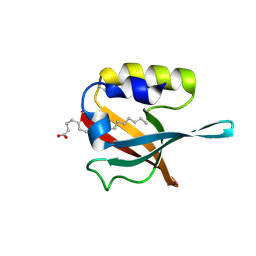 | | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid | | Descriptor: | Pullulanase C protein, VACCENIC ACID | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid
To Be Published
|
|
5DVY
 
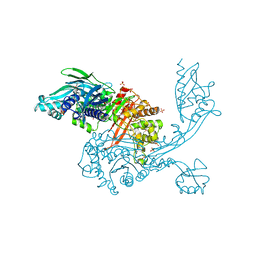 | | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Penicillin binding protein 2 prime, SULFATE ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of the Dimeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
5HW2
 
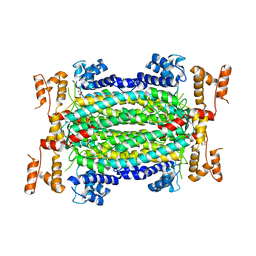 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate lyase, FUMARIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid
To Be Published
|
|
5HM8
 
 | | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD. | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenosine and NAD.
To Be Published
|
|
5HD6
 
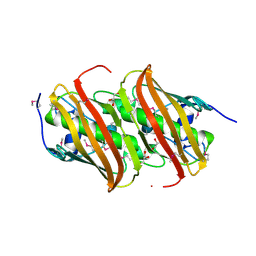 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
5EQC
 
 | | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site
To Be Published
|
|
5HJ5
 
 | | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, ACETIC ACID, FRUCTOSE -6-PHOSPHATE, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tertiary complex of glucosamine-6-phosphate deaminase from Vibrio cholerae with BETA-D-GLUCOSE-6-PHOSPHATE and FRUCTOSE -6-PHOSPHATE
To Be Published
|
|
5EUM
 
 | | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae. | | Descriptor: | ACETATE ION, CHLORIDE ION, Lipid A export ATP-binding/permease protein MsbA, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of ATP-binding Component of Fused Lipid Transporter Subunits of ABC superfamily from Haemophilus influenzae.
To Be Published
|
|
