4LYJ
 
 | | Crystal Structure of small molecule vinylsulfonamide 9 covalently bound to K-Ras G12C, alternative space group | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-07-31 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4LZ3
 
 | | F95H Epi-isozizaene synthase: Complex with Mg, inorganic pyrophosphate and benzyl triethyl ammonium cation | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, HYDROGENPHOSPHATE ION, ... | | Authors: | Li, R, Chou, W, Himmelberger, J.A, Litwin, K, Harris, G, Cane, D.E, Christianson, D.W. | | Deposit date: | 2013-07-31 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Reprogramming the Chemodiversity of Terpenoid Cyclization by Remolding the Active Site Contour of epi-Isozizaene Synthase.
Biochemistry, 53, 2014
|
|
4C3D
 
 | | HRSV M2-1, P422 crystal form | | Descriptor: | CADMIUM ION, MATRIX M2-1 | | Authors: | Tanner, S.J, Ariza, A, Richard, C.A, Wu, W, Trincao, J, Hiscox, J.A, Carroll, M.W, Silman, N.J, Eleouet, J.F, Edwards, T.A, Barr, J.N. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal Structure of the Essential Transcription Antiterminator M2-1 Protein of Human Respiratory Syncytial Virus and Implications of its Phosphorylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1FIW
 
 | | THREE-DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM RAM SPERMATOZOA | | Descriptor: | BETA-ACROSIN HEAVY CHAIN, BETA-ACROSIN LIGHT CHAIN, P-AMINO BENZAMIDINE, ... | | Authors: | Tranter, R, Read, J.A, Jones, R, Brady, R.L. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin.
Structure Fold.Des., 8, 2000
|
|
4LFZ
 
 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 Bound to Codon CCC-U in the Absence of Paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-06-27 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.92000055 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BPA
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with NAG-NAM-NAG-NAM tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, AMPDH2, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E, Lastochkin, E, Zhang, W, Hellman, L, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4CF4
 
 | |
4L9W
 
 | | Crystal Structure of H-Ras G12C, GMPPNP-bound | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Ostrem, J.M, Peters, U, Sos, M.L, Wells, J.A, Shokat, K.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions.
Nature, 503, 2013
|
|
4C7I
 
 | | Leismania major N-myristoyltransferase in complex with a peptidomimetic (-OH) molecule | | Descriptor: | DIMETHYL SULFOXIDE, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Olaleye, T.O, Brannigan, J.A, Goncalves, V, Roberts, S.M, Leatherbarrow, R.J, Wilkinson, A.J, Tate, E.W. | | Deposit date: | 2013-09-20 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Peptidomimetic Inhibitors of N-Myristoyltransferase from Human Malaria and Leishmaniasis Parasites.
Org.Biomol.Chem., 12, 2014
|
|
1FR4
 
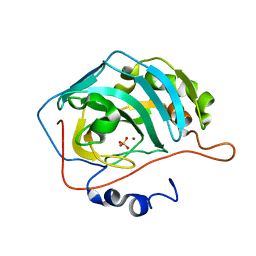 | | X-RAY CRYSTAL STRUCTURE OF COPPER-BOUND F93I/F95M/W97V CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE II, COPPER (II) ION, SULFATE ION | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-07 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
4KVD
 
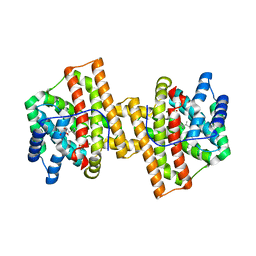 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (4aS,7S)-1,4a-dimethyl-7-(prop-1-en-2-yl)decahydroquinolin-1-ium | | Descriptor: | (1R,4aS,7S,8aR)-1,4a-dimethyl-7-(prop-1-en-2-yl)decahydroquinolinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Faraldos, J.A, Al-lami, N, Janvier, M, D'Antonio, E.L, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4DEX
 
 | |
4DFA
 
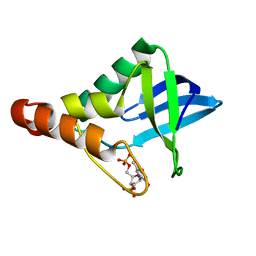 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I.A, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
4DGZ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A/L125A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Nam, S.P, Schlessman, J.L, Heroux, A, Garcia-Moreno E, B. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Pressure effects in proteins
To be Published
|
|
1YHZ
 
 | | Crystal structure of Arabidopsis thaliana Acetohydroxyacid synthase In Complex With A Sulfonylurea Herbicide, Chlorsulfuron | | Descriptor: | 1-(2-CHLOROPHENYLSULFONYL)-3-(4-METHOXY-6-METHYL-L,3,5-TRIAZIN-2-YL)UREA, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | McCourt, J.A, Pang, S.S, King-Scott, J, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2005-01-10 | | Release date: | 2006-01-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4DO7
 
 | | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2 | | Descriptor: | Amidohydrolase 2, SULFATE ION, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an amidohydrolase (cog3618) from burkholderia multivorans (target efi-500235) with bound zn, space group c2
to be published
|
|
1FSN
 
 | | X-RAY CRYSTAL STRUCTURE OF METAL-FREE F93S/F95L/W97M CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE II | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-11 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
1FFP
 
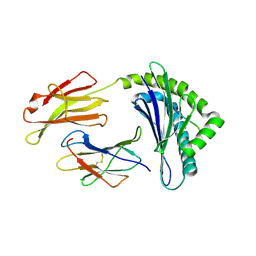 | | CRYSTAL STRUCTURE OF MURINE CLASS I H-2DB COMPLEXED WITH PEPTIDE GP33 (C9M/K1S) | | Descriptor: | BETA-2 MICROGLOBULIN BETA CHAIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B, ... | | Authors: | Wang, B, Sharma, A, Maile, R, Saad, M, Collins, E.J, Frelinger, J.A. | | Deposit date: | 2000-07-25 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Peptidic termini play a significant role in TCR recognition
J.IMMUNOL., 169, 2002
|
|
4LE8
 
 | |
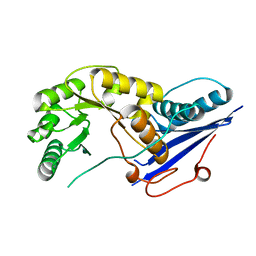
1FHU
 
 | |
1GWY
 
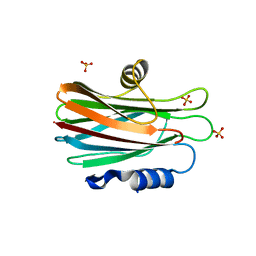 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II | | Descriptor: | STICHOLYSIN II, SULFATE ION | | Authors: | Mancheno, J.M, Martin-Benito, J, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-03-26 | | Release date: | 2003-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
4KWD
 
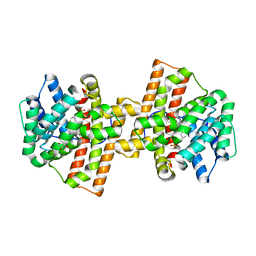 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (1R,8R,9aS)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1R,5R,8R,9aS)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4DF0
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, Orotidine 5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-22 | | Release date: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Thermoproteus neutrophilus
To be Published
|
|
4DX7
 
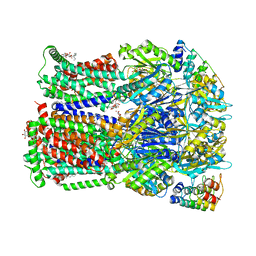 | | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop | | Descriptor: | Acriflavine resistance protein B, DARPIN, DECANE, ... | | Authors: | Eicher, T, Cha, H, Seeger, M.A, Brandstaetter, L, El-Delik, J, Bohnert, J.A, Kern, W.V, Verrey, F, Gruetter, M.G, Diederichs, K, Pos, K.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
