7CRB
 
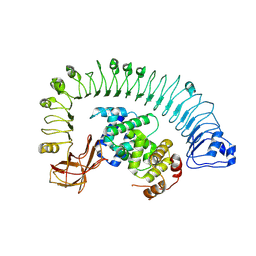 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
6P8K
 
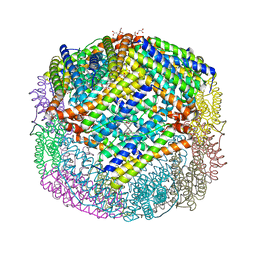 | | Escherichia coli Bacterioferritin Substituted with Zinc Protoporphyrin IX | | Descriptor: | Bacterioferritin, MALONATE ION, PROTOPORPHYRIN IX CONTAINING ZN, ... | | Authors: | Taylor, A.B, Cioloboc, D, Kurtz, D.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Zinc Porphyrin-Substituted Bacterioferritin and Photophysical Properties of Iron Reduction.
Biochemistry, 59, 2020
|
|
2X8G
 
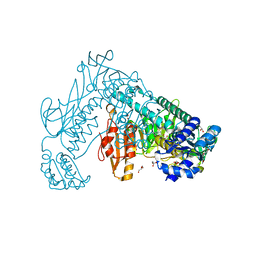 | | Oxidized thioredoxin glutathione reductase from Schistosoma mansoni | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Boumis, G, Brunori, M, Miele, A.E, Saccoccia, F, Bellelli, A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the Catalytic Cycle of Schistosoma Mansoni Thioredoxin Glutathione Reductase by X-Ray Crystallography
J.Biol.Chem., 285, 2010
|
|
3ZIM
 
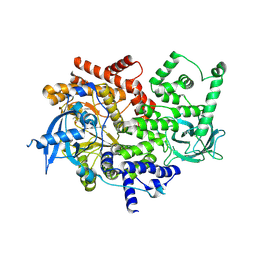 | | Discovery of a potent and isoform-selective targeted covalent inhibitor of the lipid kinase PI3Kalpha | | Descriptor: | 1-[4-[[2-(1H-indazol-4-yl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidin-6-yl]methyl]piperazin-1-yl]-6-methyl-hept-5-ene-1,4- dione, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT ALPHA ISOFORM | | Authors: | Nacht, M, Qiao, L, Sheets, M.P, Martin, T.S, Labenski, M, Mazdiyasni, H, Karp, R, Zhu, Z, Chaturvedi, P, Bhavsar, D, Niu, D, Westlin, W, Petter, R.C, Medikonda, A.P, Jestel, A, Blaesse, M, Singh, J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Potent and Isoform-Selective Targeted Covalent Inhibitor of the Lipid Kinase Pi3Kalpha
J.Med.Chem., 56, 2013
|
|
7CR1
 
 | | human KCNQ2 in complex with ztz240 | | Descriptor: | N-(6-chloranylpyridin-3-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Li, X, Lv, D, Wang, J, Ye, S, Guo, J. | | Deposit date: | 2020-08-12 | | Release date: | 2020-09-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for ligand activation of the human KCNQ2 channel.
Cell Res., 31, 2021
|
|
3ZNV
 
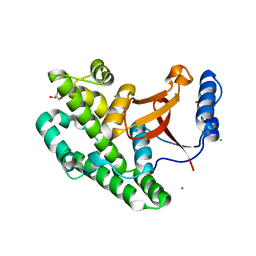 | | Crystal structure of the OTU domain of OTULIN at 1.3 Angstroms. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
6FWZ
 
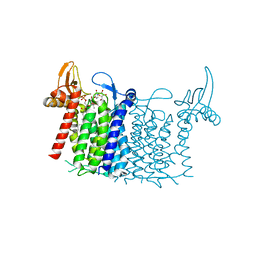 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-07 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
8PUF
 
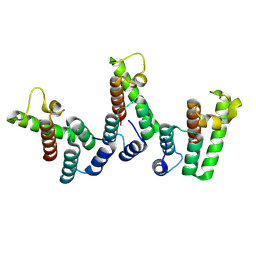 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUG
 
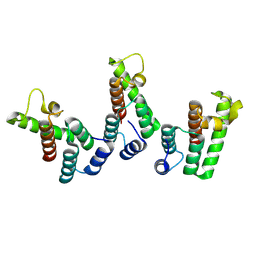 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PU6
 
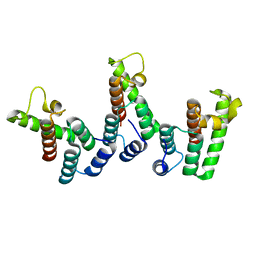 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: pooled class | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PU9
 
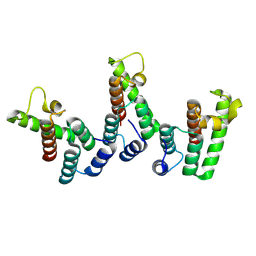 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -10 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUC
 
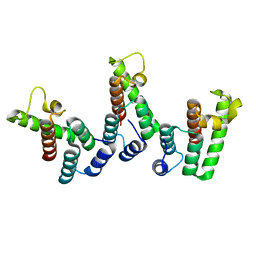 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
3HRE
 
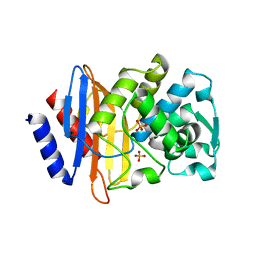 | | X-ray crystallographic structure of CTX-M-9 S70G | | Descriptor: | CTX-M-9 extended-spectrum beta-lactamase, PHOSPHATE ION | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
2X99
 
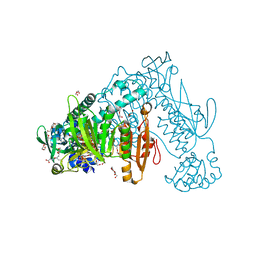 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with NADPH | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Boumis, G, Brunori, M, Miele, A.E, Saccoccia, F, Bellelli, A. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping the Catalytic Cycle of Schistosoma Mansoni Thioredoxin Glutathione Reductase by X-Ray Crystallography
J.Biol.Chem., 285, 2010
|
|
5OD3
 
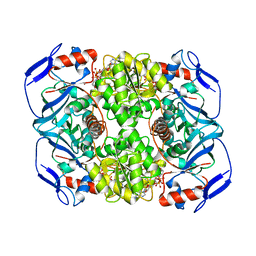 | | Crystal structure of R. ruber ADH-A, mutant Y54G, L119Y | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereo- and Regioselectivity in Catalyzed Transformation of a 1,2-Disubstituted Vicinal Diol and the Corresponding Diketone by Wild Type and Laboratory Evolved Alcohol Dehydrogenases
Acs Catalysis, 8, 2018
|
|
4B3S
 
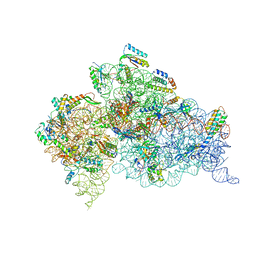 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
2AYQ
 
 | |
6RW8
 
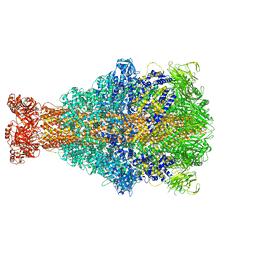 | | Cryo-EM structure of Xenorhabdus nematophila XptA1 | | Descriptor: | A component of insecticidal toxin complex (Tc) | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
6S0L
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
1WOG
 
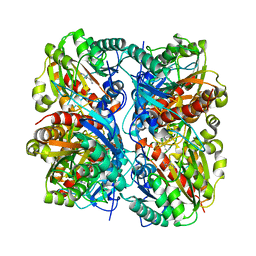 | | Crystal Structure of Agmatinase Reveals Structural Conservation and Inhibition Mechanism of the Ureohydrolase Superfamily | | Descriptor: | HEXANE-1,6-DIAMINE, MANGANESE (II) ION, agmatinase | | Authors: | Ahn, H.J, Kim, K.H, Lee, J, Ha, J.-Y, Lee, H.H, Kim, D, Yoon, H.-J, Kwon, A.-R, Suh, S.W. | | Deposit date: | 2004-08-18 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of agmatinase reveals structural conservation and inhibition mechanism of the ureohydrolase superfamily
J.Biol.Chem., 279, 2004
|
|
3DLQ
 
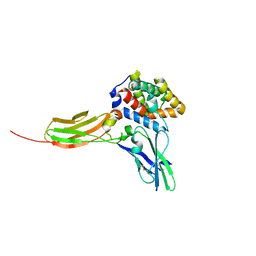 | | Crystal structure of the IL-22/IL-22R1 complex | | Descriptor: | Interleukin-22, Interleukin-22 receptor subunit alpha-1 | | Authors: | Bleicher, L, de Moura, P.R, Watanabe, L, Colau, D, Dumoutier, L, Renauld, J.-C, Polikarpov, I. | | Deposit date: | 2008-06-28 | | Release date: | 2008-08-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism
Febs Lett., 582, 2008
|
|
6RWA
 
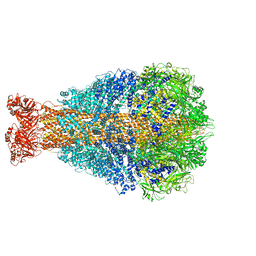 | | Cryo-EM structure of Photorhabdus luminescens TcdA4 | | Descriptor: | TcdA4 | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
4BEV
 
 | | ATPase crystal structure with bound phosphate analogue | | Descriptor: | COPPER EFFLUX ATPASE, MAGNESIUM ION, TRIFLUOROMAGNESATE | | Authors: | Mattle, D, Drachmann, N.D, Liu, X.Y, Gourdon, P, Pedersen, B.P, Morth, P, Wang, J, Nissen, P. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | ATPase Crystal Structure with Bound Phosphate Analogue
To be Published
|
|
7YHN
 
 | | ANTI-TUMOR AGENT Y48 IN COMPLEX WITH TUBULIN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methyl-3-[(4-methylphenyl)sulfonylamino]-~{N}-[(6-methylpyridin-3-yl)methyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Du, T, Ji, M, Hou, Z, Lin, S, Zhang, J, Wu, D, Zhang, K, Lu, D, Xu, H, Chen, X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Benzamide Derivatives as Potent and Orally Active Tubulin Inhibitors Targeting the Colchicine Binding Site.
J.Med.Chem., 65, 2022
|
|
6FM9
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
