4J09
 
 | | Crystal Structure of LpxA bound to RJPXD33 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Jenkins, R.J, Meagher, J.L, Stuckey, J.A, Dotson, G.D. | | Deposit date: | 2013-01-30 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Recognition of Peptide RJPXD33 by Acyltransferases in Lipid A Biosynthesis.
J.Biol.Chem., 289, 2014
|
|
4JB3
 
 | | Crystal structure of BT_0970, a had family phosphatase from bacteroides thetaiotaomicron VPI-5482, TARGET EFI-501083, with bound sodium and glycerol, closed lid, ordered loop | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Kumar, P.R, Ghosh, A, Al Obaidi, N.F, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bt_0970, a had family phosphatase from bacteroides thetaiotaomicron VPI-5482, TARGET EFI-501083, with bound sodium and glycerol, closed lid, ordered loop
To be Published
|
|
4JG0
 
 | | Structure of phosphoserine/threonine (pSTAb) scaffold bound to pSer peptide | | Descriptor: | Fab heavy chain, Fab light chain, PROPANOIC ACID, ... | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
4JCI
 
 | | Crystal structure of csal_2705, a putative hydroxyproline epimerase from CHROMOHALOBACTER SALEXIGENS (TARGET EFI-506486), SPACE GROUP P212121, unliganded | | Descriptor: | Proline racemase, SODIUM ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-21 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of csal_2705, a putative hydroxyproline epimerase from CHROMOHALOBACTER SALEXIGENS (TARGET EFI-506486), space group P212121, unliganded
To be Published
|
|
4JR2
 
 | |
1R6W
 
 | | Crystal structure of the K133R mutant of o-Succinylbenzoate synthase (OSBS) from Escherichia coli. Complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, MAGNESIUM ION, o-Succinylbenzoate Synthase | | Authors: | Klenchin, V.A, Taylor Ringia, E.A, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-10-17 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Evolution of Enzymatic Activity in the Enolase Superfamily: Structural and Mutagenic Studies of the Mechanism of the Reaction Catalyzed by o-Succinylbenzoate Synthase from Escherichia coli
Biochemistry, 42, 2003
|
|
1R9G
 
 | | Three-dimensional Structure of YaaE from Bacillus subtilis | | Descriptor: | Hypothetical protein yaaE | | Authors: | Bauer, J.A, Bennett, E.M, Begley, T.P, Ealick, S.E. | | Deposit date: | 2003-10-29 | | Release date: | 2004-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional Structure of YaaE from Bacillus subtilis, a Glutaminase Implicated in Pyridoxal-5'-phosphate Biosynthesis.
J.Biol.Chem., 279, 2004
|
|
4JPY
 
 | |
4JBB
 
 | | Crystal structure of Glutathione S-transferase A6TBY7(Target EFI-507184) from Klebsiella pneumoniae MGH 78578, GSH complex | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLUTATHIONE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase A6Tby7 (Target Efi-507184) from Klebsiella Pneumoniae
To be Published
|
|
4JFZ
 
 | | Structure of phosphoserine (pSAb) scaffold bound to pSer peptide | | Descriptor: | Fab heavy chain, Fab light chain, PROPANOIC ACID, ... | | Authors: | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
4JUU
 
 | | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand | | Descriptor: | CHLORIDE ION, Epimerase, PHOSPHATE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative hydroxyproline epimerase from xanthomonas campestris (TARGET EFI-506516) with bound phosphate and unknown ligand
To be Published
|
|
1RQ2
 
 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH CITRATE | | Descriptor: | CITRIC ACID, Cell division protein ftsZ | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
4JXK
 
 | | Crystal Structure of Oxidoreductase ROP_24000 (Target EFI-506400) from Rhodococcus opacus B4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, oxidoreductase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Oxidoreductase Rop_24000 (Target EFI-506400) from Rhodococcus Opacus
To be Published
|
|
4JXY
 
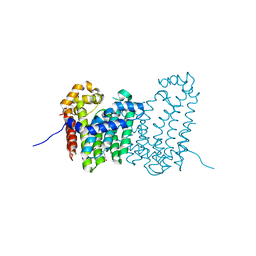 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE PATL_3739 (TARGET EFI-509195) FROM Pseudoalteromonas atlantica | | Descriptor: | GLUTARIC ACID, PHOSPHATE ION, Trans-hexaprenyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-28 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF ISOPRENOID SYNTHASE PATL_3739 FROM Pseudoalteromonas atlantica
To be Published
|
|
1RTX
 
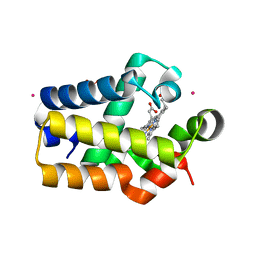 | | Crystal Structure of Synechocystis Hemoglobin with a Covalent Heme Linkage | | Descriptor: | CADMIUM ION, Cyanoglobin, POTASSIUM ION, ... | | Authors: | Hoy, J.A, Kundu, S, Trent, J.T, Ramaswamy, S, Hargrove, M.S. | | Deposit date: | 2003-12-10 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of synechocystis hemoglobin with a covalent heme linkage.
J.Biol.Chem., 279, 2004
|
|
4J9W
 
 | | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with the inhibitor pyrrole-2-carboxylate | | Descriptor: | GLYCEROL, PYRROLE-2-CARBOXYLATE, Proline racemase family protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the complex of a hydroxyproline epimerase (TARGET EFI-506499, PSEUDOMONAS FLUORESCENS PF-5) with the inhibitor pyrrole-2-carboxylate
To be Published
|
|
4JBD
 
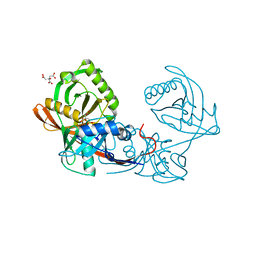 | | Crystal structure of Pput_1285, a putative hydroxyproline epimerase from Pseudomonas putida f1 (target EFI-506500), open form, space group I2, bound citrate | | Descriptor: | CITRIC ACID, Proline racemase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of pput_1285, a putative hydroxyproline epimerase from pseudomonas putida f1 (target efi-506500), open form, space group i2, bound citrate
To be Published
|
|
4JPX
 
 | |
1TYF
 
 | |
1RMX
 
 | | A SHORT LEXITROPSIN THAT RECOGNIZES THE DNA MINOR GROOVE AT 5'-ACTAGT-3': UNDERSTANDING THE ROLE OF ISOPROPYL-THIAZOLE | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3', N-[3-(DIMETHYLAMINO)PROPYL]-2-({[4-({[4-(FORMYLAMINO)-1-METHYL-1H-PYRROL-2-YL]CARBONYL}AMINO)-1-METHYL-1H-PYRROL-2-YL]CARBONYL}AMINO)-5-ISOPROPYL-1,3-THIAZOLE-4-CARBOXAMIDE | | Authors: | Anthony, N.G, Johnston, B.F, Khalaf, A.I, Mackay, S.P, Parkinson, J.A, Suckling, C.J, Waigh, R.D. | | Deposit date: | 2003-11-28 | | Release date: | 2004-08-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Short Lexitropsin that Recognizes the DNA Minor Groove at 5'-ACTAGT-3': Understanding the Role of Isopropyl-thiazole.
J.Am.Chem.Soc., 126, 2004
|
|
4GXW
 
 | | Crystal structure of a cog1816 amidohydrolase (target EFI-505188) from Burkhoderia ambifaria, with bound Zn | | Descriptor: | Adenosine deaminase, CHLORIDE ION, ZINC ION | | Authors: | Vetting, M.W, Goble, A.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a cog1816 amidohydrolase (target EFI-505188) from Burkhoderia ambifaria, with bound Zn
To be Published
|
|
4JN8
 
 | | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ENOLASE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an enolase (putative galactarate dehydratase, target efi-500740) from agrobacterium radiobacter, bound sulfate, no metal ion, ordered active site
To be Published
|
|
4JR0
 
 | | Human procaspase-3 bound to Ac-DEVD-CMK | | Descriptor: | Ac-DEVD-CMK, CHLORIDE ION, Procaspase-3 | | Authors: | Thomsen, N.D, Wells, J.A. | | Deposit date: | 2013-03-20 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1THK
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-06-01 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
4JN7
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Bouvier, J.T, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-03-14 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE (PUTATIVE GALACTARATE DEHYDRATASE, TARGET EFI-500740) FROM AGROBACTERIUM RADIOBACTER, BOUND NA and L-MALATE, ORDERED ACTIVE SITE
To be Published
|
|
