7AIK
 
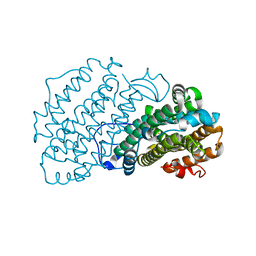 | | Ribonucleotide Reductase R2 protein from Aquifex aeolicus | | Descriptor: | FE (II) ION, Ribonucleoside-diphosphate reductase subunit beta,Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-27 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
7A5O
 
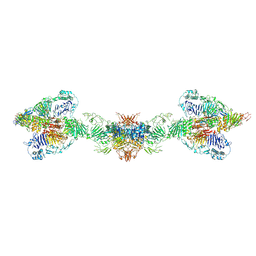 | | Human MUC2 AAs 21-1397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Khmelnitsky, L, Albert, L, Elad, N, Ilani, T, Diskin, R, Fass, D. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
1SPX
 
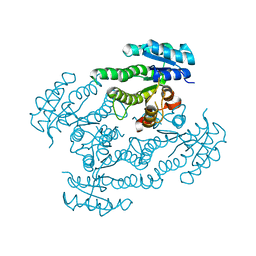 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
4WQ6
 
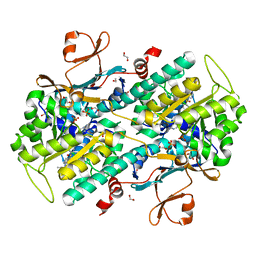 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Li, D, Wang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4FL2
 
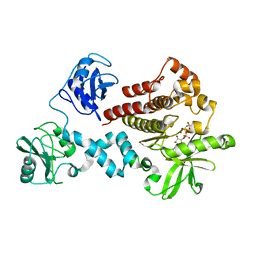 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
2P4E
 
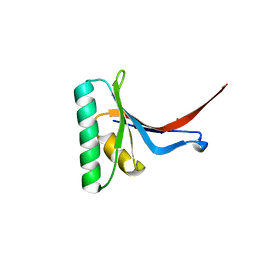 | | Crystal Structure of PCSK9 | | Descriptor: | MERCURY (II) ION, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Cunningham, D, Danley, D.E, Geoghegan, F.K, Griffor, M.C, Hawkins, J.L, Qiu, X. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7RA7
 
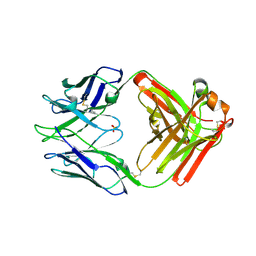 | | Crystal structure of rabbit anti-HIV Fab 11A | | Descriptor: | 11A Fab heavy chain, 11A Fab light chain, PENTAETHYLENE GLYCOL, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Limited breadth of anti-HIV Env glycan hole antibodies is further hindered by strain-specific peptide interactions
To Be Published
|
|
8VYU
 
 | | Cryo-EM Structure of the BRAF WT monomer bound to PLX8394 | | Descriptor: | (3S)-N-{3-[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl}-3-fluoropyrrolidine-1-sulfonamide, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
8VYV
 
 | | Cryo-EM Structure of the BRAF K601E monomer | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (5.86 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
4YET
 
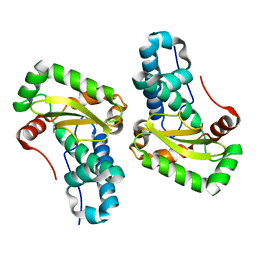 | | X-ray crystal structure of superoxide dismutase from Babesia bovis solved by Sulfur SAD | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Fairman, J.W, Clifton, M.C, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2015-02-24 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Iron superoxide dismutases in eukaryotic pathogens: new insights from Apicomplexa and Trypanosoma structures.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3TCP
 
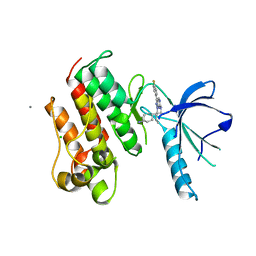 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
4YN0
 
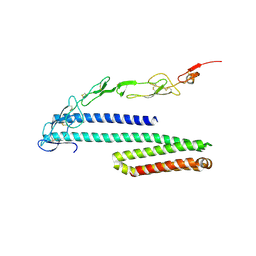 | | Crystal structure of APP E2 domain in complex with DR6 CRD domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amyloid beta A4 protein, MAGNESIUM ION, ... | | Authors: | Xu, K, Nikolov, D. | | Deposit date: | 2015-03-08 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of DR6 in complex with the amyloid precursor protein provides insight into death receptor activation.
Genes Dev., 29, 2015
|
|
1R19
 
 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (Apo structure) | | Descriptor: | fibrinogen-binding protein SdrG | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
4WTM
 
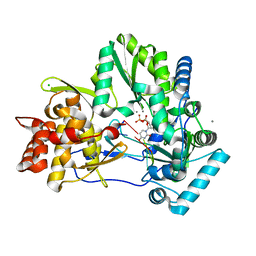 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-UAGG, RNA PRIMER 5'-PCC, MN2+, AND UDP | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, RNA PRIMER CC, ... | | Authors: | Edwards, T.E, Appleby, T.C, Fox III, D. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
1R42
 
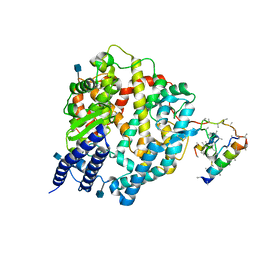 | | Native Human Angiotensin Converting Enzyme-Related Carboxypeptidase (ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ZINC ION, ... | | Authors: | Towler, P, Staker, B, Prasad, S.G, Menon, S, Ryan, D, Tang, J, Parsons, T, Fisher, M, Williams, D, Dales, N.A, Patane, M.A, Pantoliano, M.W. | | Deposit date: | 2003-10-07 | | Release date: | 2004-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis.
J.Biol.Chem., 279, 2004
|
|
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
4WZB
 
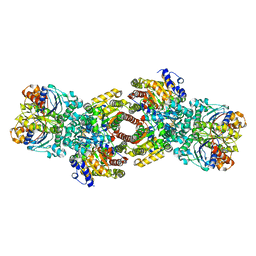 | | Crystal Structure of MgAMPPCP-bound Av2-Av1 complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nitrogenase complexes: multiple docking sites for a nucleotide switch protein.
Science, 309, 2005
|
|
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | 1.94 angstrom structure of synthetic alpha-synuclein fibrils seeding MSA neuropathology
Biorxiv, 2024
|
|
1PZ3
 
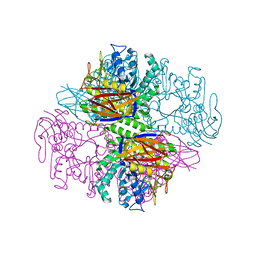 | | Crystal structure of a family 51 (GH51) alpha-L-arabinofuranosidase from Geobacillus stearothermophilus T6 | | Descriptor: | Alpha-L-arabinofuranosidase, GLYCEROL | | Authors: | Hoevel, K, Shallom, D, Niefind, K, Belakhov, V, Shoham, G, Baasov, T, Shoham, Y, Schomburg, D. | | Deposit date: | 2003-07-09 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and snapshots along the reaction pathway of a family 51 alpha-L-arabinofuranosidase
Embo J., 22, 2003
|
|
6QMT
 
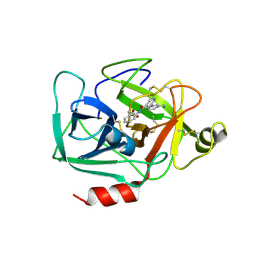 | | Complement factor D in complex with the inhibitor 2-(2-(3'-(aminomethyl)-[1,1'-biphenyl]-3-carboxamido)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
5X02
 
 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor FF-10101 | | Descriptor: | N-[(2S)-1-[5-[2-[(4-cyanophenyl)amino]-4-(propylamino)pyrimidin-5-yl]pent-4-ynylamino]-1-oxidanylidene-propan-2-yl]-4-(dimethylamino)-N-methyl-but-2-enamide, Receptor-type tyrosine-protein kinase FLT3, SULFATE ION | | Authors: | Fujikawa, N, Hirano, D, Takasaki, M, Terada, D, Hagiwara, S, Park, S.-Y, Sugiyama, K. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | A novel irreversible FLT3 inhibitor, FF-10101, shows excellent efficacy against AML cells withFLT3mutations.
Blood, 131, 2018
|
|
5V2O
 
 | | De Novo Design of Novel Covalent Constrained Meso-size Peptide Scaffolds with Unique Tertiary Structures | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Dang, B, Wu, H, Mulligan, V.K, Mravic, M, Wu, Y, Lemmin, T, Ford, A, Silva, D, Baker, D, DeGrado, W.F. | | Deposit date: | 2017-03-06 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | De novo design of covalently constrained mesosize protein scaffolds with unique tertiary structures.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8OV4
 
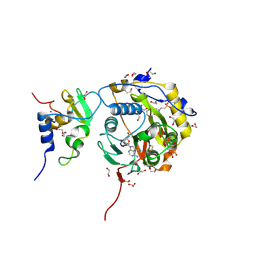 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8OV3
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
