1JQE
 
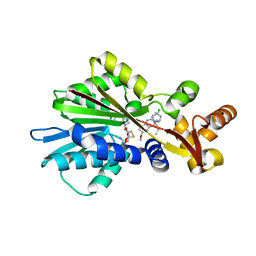 | | Crystal Structure Analysis of Human Histamine Methyltransferase (Ile105 Polymorphic Variant) Complexed with AdoHcy and Antimalarial Drug Quinacrine | | Descriptor: | Histamine N-Methyltransferase, QUINACRINE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Horton, J.R, Sawada, K, Nishibori, M, Zhang, X, Cheng, X. | | Deposit date: | 2001-08-06 | | Release date: | 2002-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons.
Structure, 9, 2001
|
|
1KHC
 
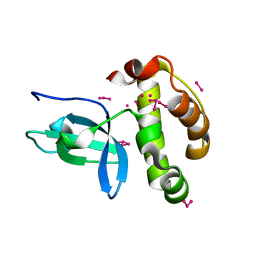 | | Crystal Structure of the PWWP Domain of Mammalian DNA Methyltransferase Dnmt3b | | Descriptor: | DNA cytosine-5 methyltransferase 3B2, UNKNOWN ATOM OR ION | | Authors: | Qiu, C, Sawada, K, Zhang, X, Cheng, X. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds.
Nat.Struct.Biol., 9, 2002
|
|
1NGN
 
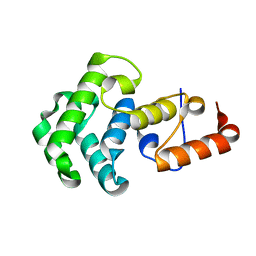 | | Mismatch repair in methylated DNA. Structure of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4 | | Descriptor: | methyl-CpG binding protein MBD4 | | Authors: | Wu, P, Qiu, C, Sohail, A, Zhang, X, Bhagwat, A.S, Cheng, X. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4
J.Biol.Chem., 278, 2003
|
|
1MVX
 
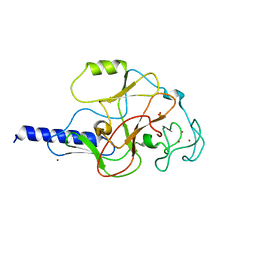 | | structure of the SET domain histone lysine methyltransferase Clr4 | | Descriptor: | CRYPTIC LOCI REGULATOR 4, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | Deposit date: | 2002-09-26 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
1MVH
 
 | | structure of the SET domain histone lysine methyltransferase Clr4 | | Descriptor: | Cryptic loci regulator 4, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
6S8N
 
 | | Cryo-EM structure of LptB2FGC in complex with lipopolysaccharide | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, Inner membrane protein yjgQ, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
6S8H
 
 | | Cryo-EM structure of LptB2FG in complex with LPS | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-5-[(2~{S},3~{S},4~{R},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{R},5~{S},6~{R})-6-[(1~{S})-2-[(2~{S},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-1-oxidanyl-ethyl]-3,4-bis(oxidanyl)-5-phosphonooxy-oxan-2-yl]oxy-3-oxidanyl-5-phosphonooxy-oxan-2-yl]oxy-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
1DP4
 
 | | DIMERIZED HORMONE BINDING DOMAIN OF THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATRIAL NATRIURETIC PEPTIDE RECEPTOR A, ... | | Authors: | van den Akker, F, Zhang, X, Miyagi, M, Huo, X, Misono, K.S, Yee, V.C. | | Deposit date: | 1999-12-23 | | Release date: | 2000-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor.
Nature, 406, 2000
|
|
5K9N
 
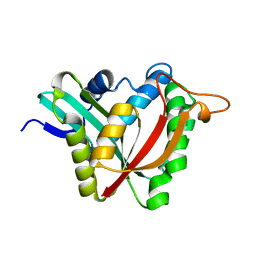 | | Structural and Mechanistic Analysis of Drosophila melanogaster Polyamine N acetyltransferase, an enzyme that Catalyzes the Formation of N acetylagmatine | | Descriptor: | Polyamine N acetyltransferase | | Authors: | Dempsey, D.R, Nichols, D.A, Battistini, M.R, Pemberton, O, Ospina, S.R, Zhang, X, Carpenter, A.-M, Chen, Y, Merkler, D.J. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Analysis of Drosophila melanogaster Agmatine N-Acetyltransferase, an Enzyme that Catalyzes the Formation of N-Acetylagmatine.
Sci Rep, 7, 2017
|
|
5OQ4
 
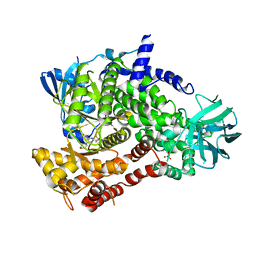 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
5OAF
 
 | | Human Rvb1/Rvb2 heterohexamer in INO80 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Aramayo, R.J, Bythell-Douglas, R, Ayala, R, Willhoft, O, Wigley, D, Zhang, X. | | Deposit date: | 2017-06-21 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM structures of the human INO80 chromatin-remodeling complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6S8G
 
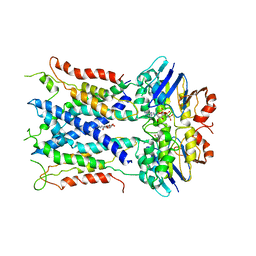 | | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | | Descriptor: | Inner membrane protein yjgQ, LPS export ABC transporter permease LptF, Lipopolysaccharide ABC transporter, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
2R5W
 
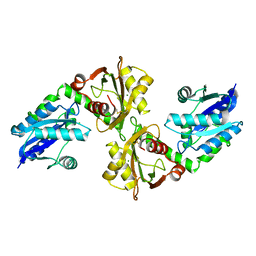 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
8R64
 
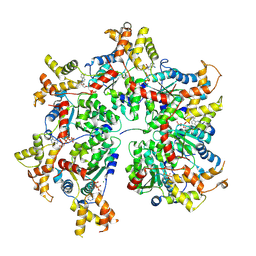 | | Cryo-EM structure of the FIGNL1 AAA hexamer bound to RAD51 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 1, ... | | Authors: | Carver, A, Yates, L.A, Zhang, X. | | Deposit date: | 2023-11-20 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of FIGNL1 in dissociating RAD51 from DNA and chromatin.
Science, 387, 2025
|
|
8QN4
 
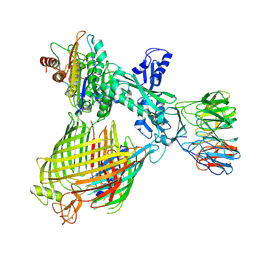 | | Structure of BAM-EspP complex in the non-closing EspP state | | Descriptor: | EspP epsilon, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Xie, T, Pang, J, Shen, C, Chang, S, Tang, X, Zhang, X, Dong, H, Zhou, R. | | Deposit date: | 2023-09-25 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Dynamic topology-mediated maturation of beta-barrel proteins in BAM-catalyzed folding
To Be Published
|
|
8BNZ
 
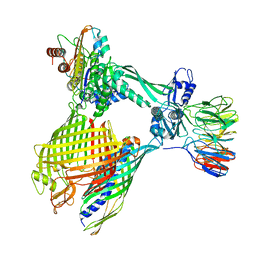 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BO2
 
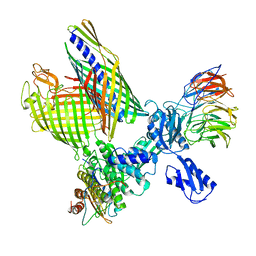 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8RE4
 
 | |
8REE
 
 | |
8RED
 
 | |
8REB
 
 | |
8REC
 
 | |
8REA
 
 | |
8QOT
 
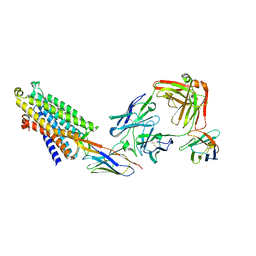 | | Structure of the mu opioid receptor bound to the antagonist nanobody NbE | | Descriptor: | Anti-Fab Nanobody, Mu-type opioid receptor, NabFab HC, ... | | Authors: | Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Manglik, A, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of mu-opioid receptor targeting by a nanobody antagonist.
Nat Commun, 15, 2024
|
|
9JF4
 
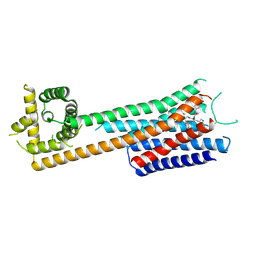 | | cryo-EM structure of Neuromedin B receptor in complex with PD168368 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-methyl-2-[(4-nitrophenyl)carbamoylamino]-~{N}-[(1-pyridin-2-ylcyclohexyl)methyl]propanamide, Neuromedin-B receptor,de novo design protein | | Authors: | Gao, K, Zhang, X, Liu, X. | | Deposit date: | 2024-09-03 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | cryo-EM structure of Neuromedin B receptor in complex with PD168368
To Be Published
|
|
