6IW7
 
 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | (2S)-2-benzamido-4-methyl-pentanoic acid, 4-[(4-fluoro-2-methyl-3H-indol-5-yl)oxy]-6-methoxy-7-[3-(pyrrolidin-1-yl)propoxy]quinazoline, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2018-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.692121 Å) | | Cite: | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease
To Be Published
|
|
7Y8K
 
 | | Structure of ScIRED wild-type from Streptomyces clavuligerus | | Descriptor: | Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7Y8N
 
 | | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(2,5-difluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 5-[2,5-bis(fluoranyl)phenyl]-3,4-dihydro-2~{H}-pyrrole, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
7Y9M
 
 | | Crystal structure of P450 BM3-2F from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of P450 BM3-2F from Bacillus megaterium
to be published
|
|
7Y9L
 
 | | Crystal structure of P450 BM3-2F from Bacillus megaterium in complex with 2-Hydroxy-5-Nitrobenzonitrile | | Descriptor: | 5-nitro-2-oxidanyl-benzenecarbonitrile, Bifunctional cytochrome P450/NADPH--P450 reductase, NICKEL (II) ION, ... | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of P450 BM3-2F from Bacillus megaterium in complex with 2-Hydroxy-5-Nitrobenzonitrile
to be published
|
|
7YAO
 
 | |
4S1R
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H08.F-117225, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H08.F-117225 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1Q
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H03+06.D-001739, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H03+06.D-001739 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1S
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H5.F-185917, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab of VRC01 light chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
7YRE
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
2L37
 
 | | 3D solution structure of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical) | | Descriptor: | Ribosome-inactivating protein luffin P1 | | Authors: | Ng, Y.M, Yang, Y, Sze, K.H, Zhang, X, Zheng, Y.T, Shaw, P.C. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural characterization and anti-HIV-1 activities of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrical).
J.Struct.Biol., 2010
|
|
2LCG
 
 | | Solution NMR structure of protein Rmet_5065 from Ralstonia metallidurans, Northeast Structural Genomics Consortium Target CrR115 | | Descriptor: | Uncharacterized protein | | Authors: | Ramelot, T.A, Yang, Y, Wang, D, Ciccosanti, C, Janjua, H, Nair, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Rmet_5065 from Ralstonia metallidurans.
Northeast Structural Genomics Consortium Target CrR115.
To be Published
|
|
2L8V
 
 | | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC (20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR219A | | Descriptor: | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod | | Authors: | Ramelot, T.A, Yang, Y, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-26 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC
(20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics
Consortium Target TeR219A
To be Published
|
|
2L06
 
 | | Solution NMR structure of the PBS linker polypeptide domain (fragment 254-400) of phycobilisome linker protein ApcE from Synechocystis sp. PCC 6803. Northeast Structural Genomics Consortium Target SgR209C | | Descriptor: | Phycobilisome LCM core-membrane linker polypeptide | | Authors: | Ramelot, T.A, Yang, Y, Cort, J.R, Hamilton, K, Ciccosanti, C, Lee, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the PBS linker polypeptide domain of phycobilisome
linker protein apcE from Synechocystis sp. Northeast Structural Genomics Consortium
Target SgR209C
To be Published
|
|
2LL8
 
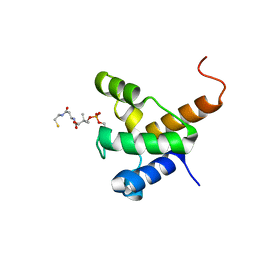 | | Solution NMR structure of the specialized holo-acyl carrier protein RPA2022 from Rhodopseudomonas palustris refined with NH RDCs, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Specialized acyl carrier protein | | Authors: | Ramelot, T.A, Ni, S, Rossi, P, Yang, Y, Wang, H, Ciccosanti, C, Maglaqui, M, Janjua, H, Nair, R, Roset, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
2L3B
 
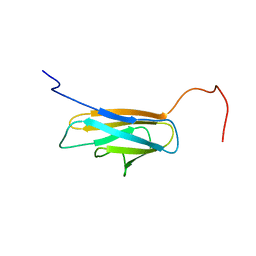 | | Solution NMR structure of the BT_0084 lipoprotein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Consortium Target BtR376 | | Descriptor: | Conserved protein found in conjugate transposon | | Authors: | Ramelot, T.A, Yang, Y, Wang, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-10 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of BT_0084, a conjugative transposon lipoprotein from Bacteroides thetaiotamicron.
Proteins, 80, 2012
|
|
2LM3
 
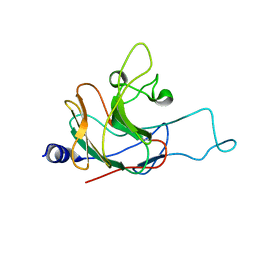 | | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevski, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D.N. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LEM
 
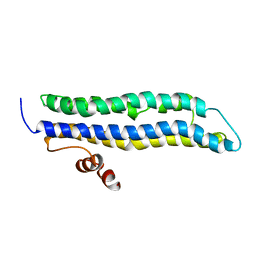 | |
2LXU
 
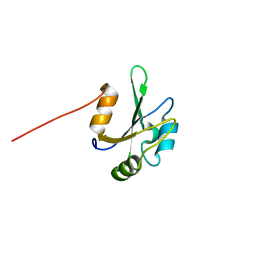 | | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A | | Descriptor: | Heterogeneous nuclear ribonucleoprotein H | | Authors: | Ramelot, T.A, Yang, Y, Pederson, K, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the eukaryotic RNA recognition motif, RRM1, from the heterogeneous nuclear ribonucleoprotein H from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8614A
To be Published
|
|
2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | Descriptor: | CalU16 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
2MA5
 
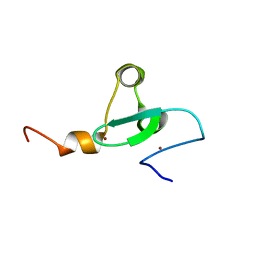 | | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7375C | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Hassan, F, Ramelot, T.A, Yang, Y, Cort, J.R, Janjua, H, Kohan, E, Lee, D, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of PHD type Zinc finger domain of Lysine-specific
demethylase 5B (PLU-1/JARID1B) from Homo sapiens, Northeast Structural
Genomics Consortium (NESG) Target HR7375C
To be Published
|
|
2MPV
 
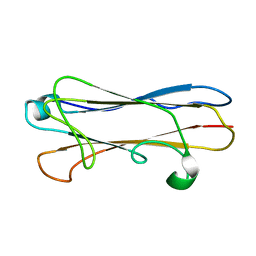 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
5KWQ
 
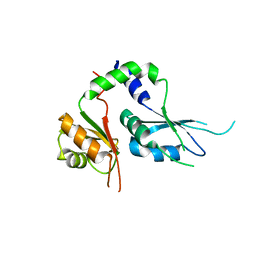 | | Two Tandem RRM Domains of FBP-Interacting Repressor (FIR), also Known as PUF60 | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Yang, Y, Zhou, H, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-18 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
2LPK
 
 | | Solution NMR of the specialized apo-acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR324 | | Descriptor: | Specialized acyl carrier protein | | Authors: | Ramelot, T.A, Rossi, P, Yang, Y, Lee, H, Ertekin, A, Wang, H, Ciccosanti, C, Kohan, E, Maglaqui, M, Janjua, H, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-14 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a specialized acyl carrier protein essential for lipid A biosynthesis with very long-chain fatty acids in open and closed conformations.
Biochemistry, 51, 2012
|
|
2OS6
 
 | |
