4EN2
 
 | |
5HH5
 
 | |
4EMZ
 
 | |
6YYN
 
 | | Structure of Cathepsin S in complex with Compound 14 | | Descriptor: | CITRATE ANION, Cathepsin S, SULFATE ION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYO
 
 | | Structure of Cathepsin S in complex with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylsulfonylpiperazin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYP
 
 | | Structure of Cathepsin S in complex with Compound 2 | | Descriptor: | 1-(furan-2-ylmethyl)-5-(trifluoromethyl)benzimidazol-2-amine, ACETATE ION, Cathepsin S, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYQ
 
 | | Structure of Cathepsin S in complex with Compound 3 | | Descriptor: | (6~{R})-2-phenyl-5,6,7,8-tetrahydroquinazolin-6-amine, Cathepsin S | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YYR
 
 | | Structure of Cathepsin S in complex with Compound 20b | | Descriptor: | (2~{R})-~{N}-(2-azanylideneethyl)-2-[2-(3-methyl-1,2-oxazol-5-yl)ethanoylamino]-3-(4-pyridin-2-ylpiperazin-1-yl)sulfonyl-propanamide, 1,2-ETHANEDIOL, CITRATE ANION, ... | | Authors: | Wagener, M, Schade, M, Merla, B, Hars, U, Kueckelhaus, S.Q. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Highly Selective Sub-Nanomolar Cathepsin S Inhibitors by Merging Fragment Binders with Nitrile Inhibitors.
J.Med.Chem., 63, 2020
|
|
4ZPL
 
 | | Crystal Structure of Protocadherin Beta 1 EC1-3 | | Descriptor: | CALCIUM ION, Protein Pcdhb1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goodman, K.M, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZPN
 
 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 with extended N-terminus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Goodman, K.M, Wolcott, H.N, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
1GFZ
 
 | | FLAVOPIRIDOL INHIBITS GLYCOGEN PHOSPHORYLASE BY BINDING AT THE INHIBITOR SITE | | Descriptor: | CAFFEINE, GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-06-29 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site.
J.Biol.Chem., 275, 2000
|
|
1FUJ
 
 | | PR3 (MYELOBLASTIN) | | Descriptor: | PR3, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Fujinaga, M, Chernaia, M.M, Halenbeck, R, Koths, K, James, M.N.G. | | Deposit date: | 1996-01-25 | | Release date: | 1996-07-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of PR3, a neutrophil serine proteinase antigen of Wegener's granulomatosis antibodies.
J.Mol.Biol., 261, 1996
|
|
2L0X
 
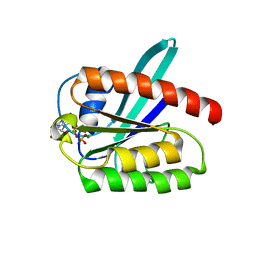 | | Solution structure of the 21 kDa GTPase RHEB bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Stoll, R, Heumann, R, Berghaus, C, Kock, G. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Ras homolog enriched in brain (Rheb) enhances apoptotic signaling.
J.Biol.Chem., 285, 2010
|
|
8WT9
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution) | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8WT7
 
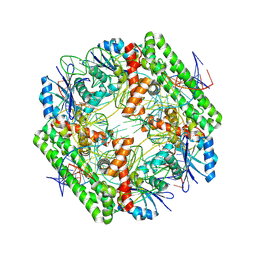 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8WT6
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange state | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8WT8
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate) | | Descriptor: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
3HDN
 
 | |
3HDM
 
 | |
1B9N
 
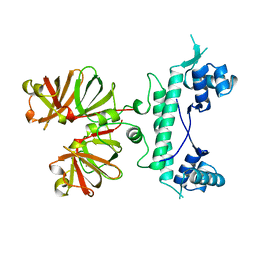 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
1B9M
 
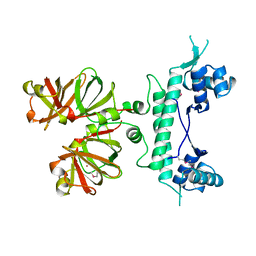 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
9EZX
 
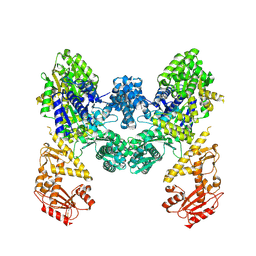 | | Vibrio cholerae DdmD apo complex | | Descriptor: | Helicase/UvrB N-terminal domain-containing protein | | Authors: | Loeff, L, Jinek, M. | | Deposit date: | 2024-04-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Molecular mechanism of plasmid elimination by the DdmDE defense system.
Science, 385, 2024
|
|
3M51
 
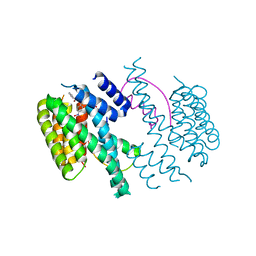 | | Structure of the 14-3-3/PMA2 complex stabilized by Pyrrolidone1 | | Descriptor: | 14-3-3-like protein C, 2-hydroxy-5-[(5S)-3-hydroxy-5-(4-nitrophenyl)-2-oxo-4-(phenylcarbonyl)-2,5-dihydro-1H-pyrrol-1-yl]benzoic acid, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Ottmann, C, Rose, R, Waldmann, H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3M50
 
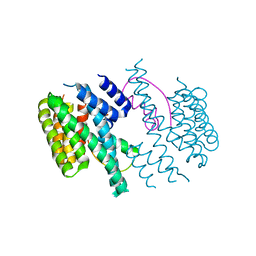 | | Structure of the 14-3-3/PMA2 complex stabilized by Epibestatin | | Descriptor: | 14-3-3-like protein C, N-[(2R,3R)-3-amino-2-hydroxy-4-phenylbutanoyl]-L-leucine, N.plumbaginifolia H+-translocating ATPase mRNA | | Authors: | Ottmann, C, Rose, R, Waldmann, H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
4XNF
 
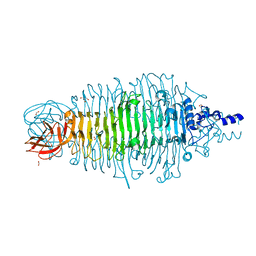 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
