7YOJ
 
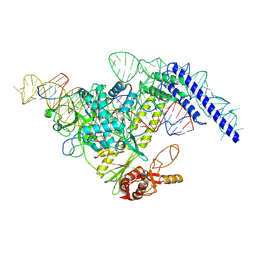 | | Structure of CasPi with guide RNA and target DNA | | Descriptor: | CasPi, DNA (30-MER), DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Li, C.P, Wang, J, Liu, J.J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The compact Cas pi (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation.
Cell Res., 33, 2023
|
|
7JVN
 
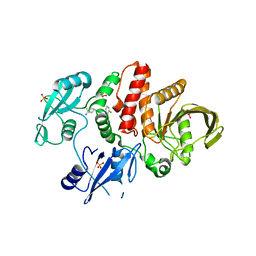 | |
7JVM
 
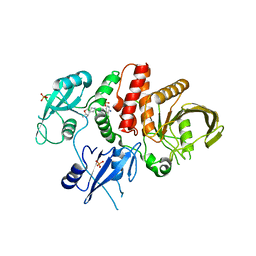 | |
6OX1
 
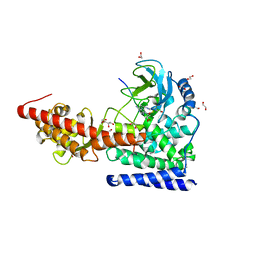 | | SETD3 in Complex with an Actin Peptide with Target Histidine Partially Methylated | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX3
 
 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX0
 
 | | SETD3 in Complex with an Actin Peptide with Sinefungin Replacing SAH as Cofactor | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Histone-lysine N-methyltransferase setd3, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX2
 
 | | SETD3in Complex with an Actin Peptide with the Target Histidine Fully Methylated | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX4
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
6OX5
 
 | | A SETD3 Mutant (N255A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, Actin-histidine N-methyltransferase, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
5UHF
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with D-IX336 | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.345 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UHG
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with D-AAP1 and Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.971 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UH6
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 2ntRNA in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.837 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UH8
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 4nt RNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.176 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UH9
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 2nt RNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.402 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UHE
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with D-AAP1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.039 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
6Y6H
 
 | | Crystal structure of STK17b (DRAK2) in complex with UNC-AP-194 probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(1-benzothiophen-2-yl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6Y6F
 
 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
5UH5
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 3 nt of RNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-10 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.746 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UH7
 
 | |
5UHC
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 3nt RNA in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*CP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UHD
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex containing 4nt RNA in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
5UHA
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.906 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
5UHB
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex in complex with Rifampin | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2017-01-11 | | Release date: | 2017-04-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (4.29 Å) | | Cite: | Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition.
Mol. Cell, 66, 2017
|
|
6ZJF
 
 | | Crystal structure of STK17B (DRAK2) in complex with AP-229 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-cyclopropylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Picado, A, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
