5CSQ
 
 | | The structure of the NK1 fragment of HGF/SF complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS9
 
 | | The structure of the NK1 fragment of HGF/SF complexed with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS1
 
 | | The structure of the NK1 fragment of HGF/SF | | Descriptor: | Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CT1
 
 | | The structure of the NK1 fragment of HGF/SF complexed with CHES | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
5CS5
 
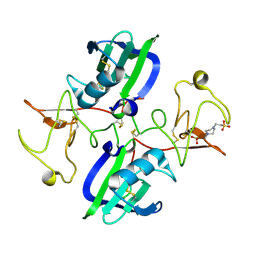 | | The structure of the NK1 fragment of HGF/SF complexed with PIPES | | Descriptor: | Hepatocyte growth factor, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
2ASU
 
 | |
2BB2
 
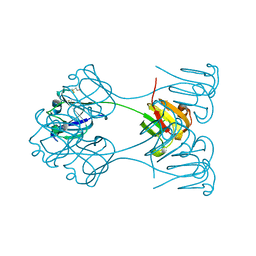 | | X-RAY ANALYSIS OF BETA B2-CRYSTALLIN AND EVOLUTION OF OLIGOMERIC LENS PROTEINS | | Descriptor: | BETA B2-CRYSTALLIN, BETA-MERCAPTOETHANOL | | Authors: | Bax, B, Lapatto, R, Nalini, V, Driessen, H, Lindley, P.F, Mahadevan, D, Blundell, T.L, Slingsby, C. | | Deposit date: | 1992-09-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray analysis of beta B2-crystallin and evolution of oligomeric lens proteins.
Nature, 347, 1990
|
|
1YON
 
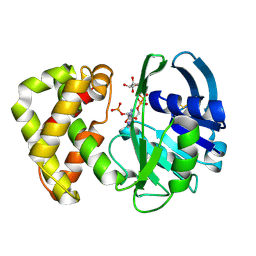 | | Escherichia coli ketopantoate reductase in complex with 2-monophosphoadenosine-5'-diphosphate | | Descriptor: | 2-dehydropantoate 2-reductase, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4R,5R)-3,4,5-TRIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Ciulli, A, Lobley, C.M.C, Tuck, K.L, Williams, G, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2005-01-28 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | pH-tuneable binding of 2'-phospho-ADP-ribose to ketopantoate reductase: a structural and calorimetric study.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1QGX
 
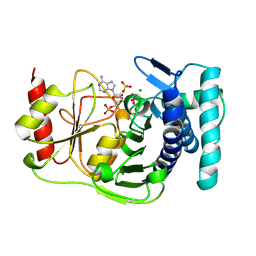 | | X-RAY STRUCTURE OF YEAST HAL2P | | Descriptor: | 3',5'-ADENOSINE BISPHOSPHATASE, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Albert, A, Yenush, L, Gil-Mascarell, M.R, Rodriguez, P.L, Patel, J, Martinez-Ripoll, M, Blundell, T.L, Serrano, R. | | Deposit date: | 1999-05-10 | | Release date: | 2000-01-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of yeast Hal2p, a major target of lithium and sodium toxicity, and identification of framework interactions determining cation sensitivity.
J.Mol.Biol., 295, 2000
|
|
1Z56
 
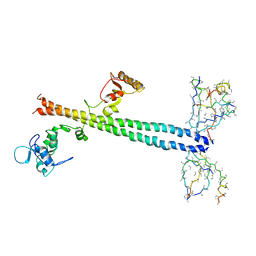 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
1YYH
 
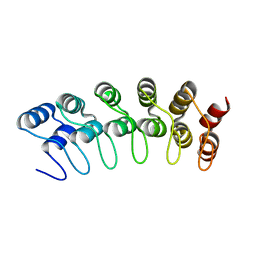 | | Crystal structure of the human Notch 1 ankyrin domain | | Descriptor: | Notch 1, ankyrin domain | | Authors: | Ehebauer, M.T, Chirgadze, D.Y, Hayward, P, Martinez-Arias, A, Blundell, T.L. | | Deposit date: | 2005-02-25 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | High-resolution crystal structure of the human Notch 1 ankyrin domain
Biochem.J., 392, 2005
|
|
2C81
 
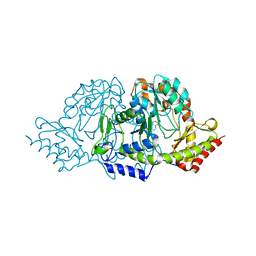 | | Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis. | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLUTAMINE-2-DEOXY-SCYLLO-INOSOSE AMINOTRANSFERASE | | Authors: | Popovic, B, Tang, X, Chirgadze, D.Y, Huang, F, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2005-11-30 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis.
Proteins, 65, 2006
|
|
2C7T
 
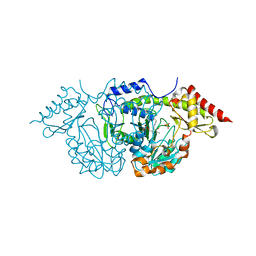 | | CRYSTAL STRUCTURE OF THE PLP-BOUND FORM OF BTRR, A DUAL FUNCTIONAL AMINOTRANSFERASE INVOLVED IN BUTIROSIN BIOSYNTHESIS. | | Descriptor: | GLUTAMINE-2-DEOXY-SCYLLO-INOSOSE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Popovic, B, Tang, X, Chirgadze, D.Y, Huang, F, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2005-11-29 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the PLP- and PMP-bound forms of BtrR, a dual functional aminotransferase involved in butirosin biosynthesis.
Proteins, 65, 2006
|
|
4H7Y
 
 | |
4KI7
 
 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinase from Mycobacterium tuberculosis - compound 41c [3-hydroxy-5-(3-nitrophenoxy)benzoic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 3-hydroxy-5-(3-nitrophenoxy)benzoic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-01 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KIJ
 
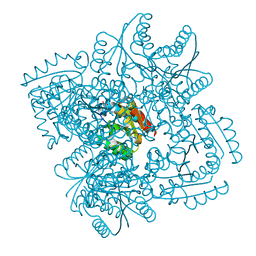 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinase dehydratase from Mycobacterium tuberculosis - compound 35c [3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid] | | Descriptor: | 3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KL9
 
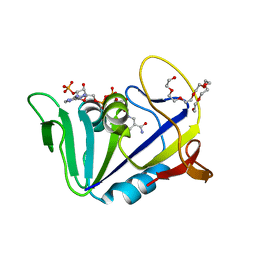 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in the space group C2 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KG0
 
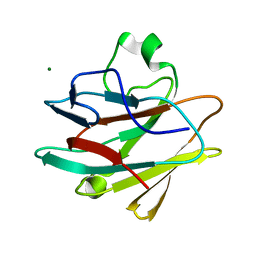 | | Crystal structure of the drosophila melanogaster neuralized-nhr1 domain | | Descriptor: | MAGNESIUM ION, Protein neuralized | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2013-04-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure, biochemical and biophysical characterisation of NHR1 domain of E3 Ubiquitin ligase neutralized
Advances in Enzyme Research, 1, 2013
|
|
4KM2
 
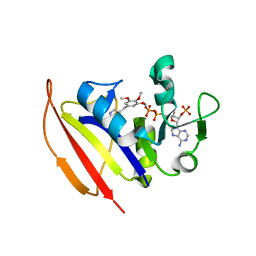 | |
4KM0
 
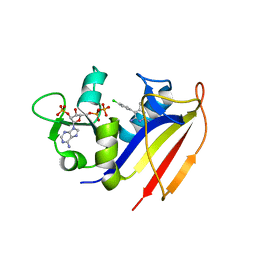 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with pyrimethamine | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KIW
 
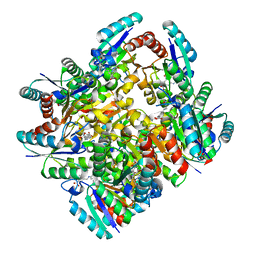 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49e [5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KLX
 
 | |
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KIU
 
 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49d [5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
1QIB
 
 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
