1BN1
 
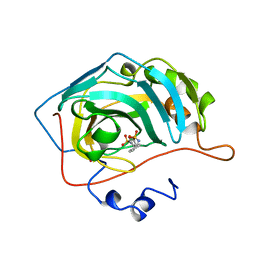 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, THIOPHENE-2,5-DISULFONIC ACID 2-AMIDE-5-(4-METHYL-BENZYLAMIDE), ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-31 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNQ
 
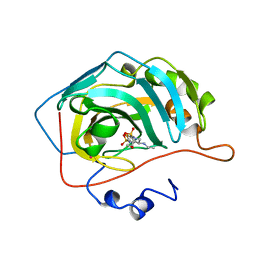 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (R)-4-ETHYLAMINO-3,4-DIHYDRO-2-(2-METHOYLETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
1BNV
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (S)-3,4-DIHYDRO-2-(3-METHOXYPHENYL)-4-METHYLAMINO-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
2LA5
 
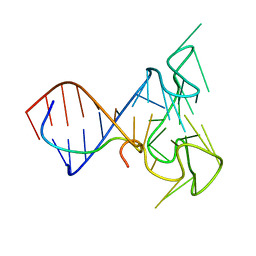 | | RNA Duplex-Quadruplex Junction Complex with FMRP RGG peptide | | Descriptor: | Fragile X mental retardation 1 protein, RNA (36-MER) | | Authors: | Phan, A, Kuryavyi, V, Darnell, J, Serganov, A, Majumdar, A, Ilin, S, Darnell, R, Patel, D. | | Deposit date: | 2011-03-03 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2JQ7
 
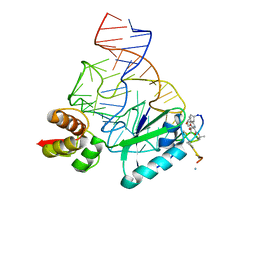 | | Model for thiostrepton binding to the ribosomal L11-RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RIBOSOMAL RNA, THIOSTREPTON | | Authors: | Jonker, H.R.A, Ilin, S, Grimm, S.K, Woehnert, J, Schwalbe, H. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | L11 Domain Rearrangement Upon Binding to RNA and Thiostrepton Studied by NMR Spectroscopy
Nucleic Acids Res., 35, 2007
|
|
1A9M
 
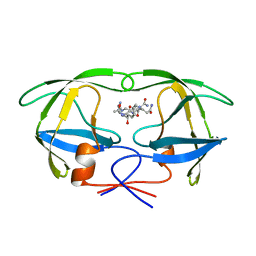 | | G48H MUTANT OF HIV-1 PROTEASE IN COMPLEX WITH A PEPTIDIC INHIBITOR U-89360E | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Zhang, X.-J, Foundling, S, Hartsuck, J.A, Tang, J. | | Deposit date: | 1998-04-08 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a G48H mutant of HIV-1 protease explains how glycine-48 replacements produce mutants resistant to inhibitor drugs.
FEBS Lett., 420, 1997
|
|
1CAM
 
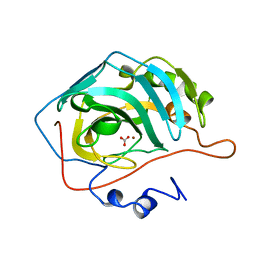 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1CAL
 
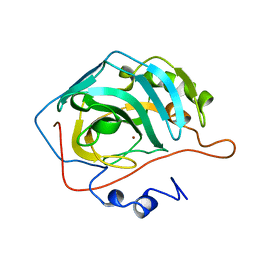 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1CAI
 
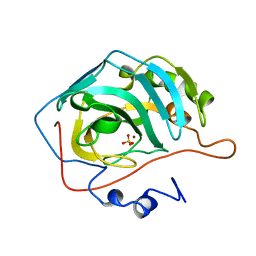 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, SULFATE ION, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1CAK
 
 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, SULFATE ION, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1CAJ
 
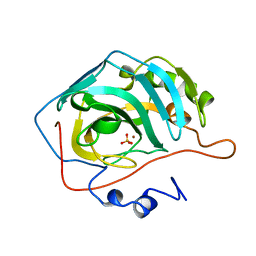 | | STRUCTURAL ANALYSIS OF THE ZINC HYDROXIDE-THR 199-GLU 106 HYDROGEN BONDING NETWORK IN HUMAN CARBONIC ANHYDRASE II | | Descriptor: | CARBONIC ANHYDRASE II, SULFATE ION, ZINC ION | | Authors: | Xue, Y, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-17 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the zinc hydroxide-Thr-199-Glu-106 hydrogen-bond network in human carbonic anhydrase II.
Proteins, 17, 1993
|
|
1AUT
 
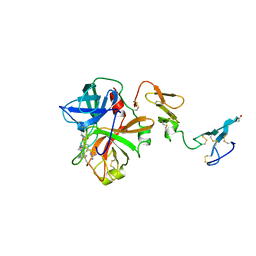 | | Human activated protein C | | Descriptor: | ACTIVATED PROTEIN C, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide | | Authors: | Mather, T, Oganessyan, V, Hof, P, Bode, W, Huber, R, Foundling, S, Esmon, C. | | Deposit date: | 1996-06-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A crystal structure of Gla-domainless activated protein C.
EMBO J., 15, 1996
|
|
1AXA
 
 | | ACTIVE-SITE MOBILITY IN HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE AS DEMONSTRATED BY CRYSTAL STRUCTURE OF A28S MUTANT | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Hartsuck, J.A, Foundling, S, Ermolieff, J, Tang, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant.
Protein Sci., 7, 1998
|
|
1BIC
 
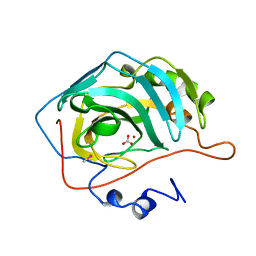 | | CRYSTALLOGRAPHIC ANALYSIS OF THR-200-> HIS HUMAN CARBONIC ANHYDRASE II AND ITS COMPLEX WITH THE SUBSTRATE, HCO3- | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Xue, Y, Vidgren, J, Svensson, L.A, Liljas, A, Jonsson, B.-H, Lindskog, S. | | Deposit date: | 1992-09-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of Thr-200-->His human carbonic anhydrase II and its complex with the substrate, HCO3-.
Proteins, 15, 1993
|
|
5NZT
 
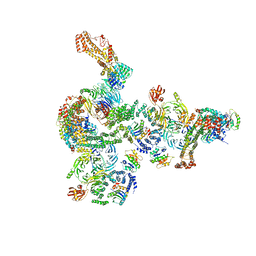 | | The structure of the COPI coat linkage I | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZV
 
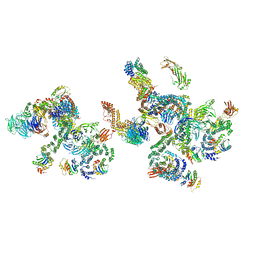 | | The structure of the COPI coat linkage IV | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (17.299999 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZU
 
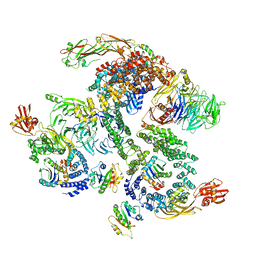 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | Authors: | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
2VT2
 
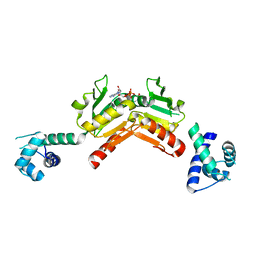 | | Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, REDOX-SENSING TRANSCRIPTIONAL REPRESSOR REX | | Authors: | Wang, E, Bauer, M.C, Rogstam, A, Linse, S, Logan, D.T, von Wachenfeldt, C. | | Deposit date: | 2008-05-08 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex.
Mol. Microbiol., 69, 2008
|
|
2VT3
 
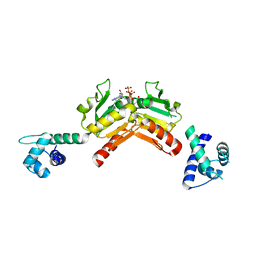 | | Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, REDOX-SENSING TRANSCRIPTIONAL REPRESSOR REX | | Authors: | Wang, E, Bauer, M.C, Rogstam, A, Linse, S, Logan, D, von Wachenfeldt, C. | | Deposit date: | 2008-05-08 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex.
Mol. Microbiol., 69, 2008
|
|
3QT0
 
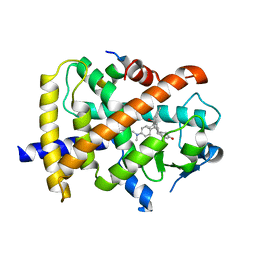 | | Revealing a steroid receptor ligand as a unique PPARgamma agonist | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Nuclear receptor coactivator 1 peptide, Peroxisome proliferator-activated receptor gamma | | Authors: | Rong, H. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Revealing a steroid receptor ligand as a unique PPAR gamma agonist.
Cell Res., 22, 2012
|
|
3NWW
 
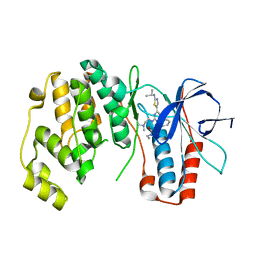 | | P38 Alpha kinase complexed with a 2-aminothiazol-5-yl-pyrimidine based inhibitor | | Descriptor: | 1-[2-(2-{[2-(dimethylamino)ethyl]amino}-6-{2-[(1-methylethyl)amino]-1,3-thiazol-5-yl}pyrimidin-4-yl)benzyl]-3-ethylurea, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Utilization of a nitrogen-sulfur nonbonding interaction in the design of new 2-aminothiazol-5-yl-pyrimidines as p38alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8WPU
 
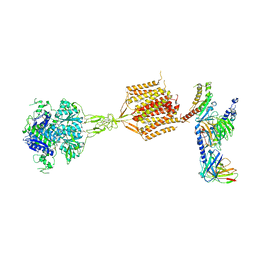 | | Human calcium-sensing receptor(CaSR) bound to cinacalcet in complex with Gq protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPG
 
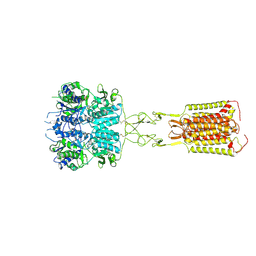 | | Human calcium-sensing receptor bound with cinacalcet in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
2POI
 
 | | Crystal structure of XIAP BIR1 domain (I222 form) | | Descriptor: | Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Lin, S. | | Deposit date: | 2007-04-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | XIAP Induces NF-kappaB Activation via the BIR1/TAB1 Interaction and BIR1 Dimerization.
Mol.Cell, 26, 2007
|
|
7EEB
 
 | | Structure of the CatSpermasome | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J.P, Ke, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a mammalian sperm cation channel complex.
Nature, 595, 2021
|
|
