3FTR
 
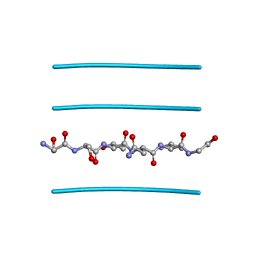 | |
3FVA
 
 | | NNQNTF segment from elk prion | | Descriptor: | Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2009-01-15 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.458 Å) | | Cite: | Molecular mechanisms for protein-encoded inheritance.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3FTK
 
 | |
4GMR
 
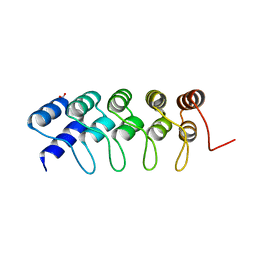 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR266. | | Descriptor: | NITRATE ION, OR266 DE NOVO PROTEIN | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Mao, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
4GPM
 
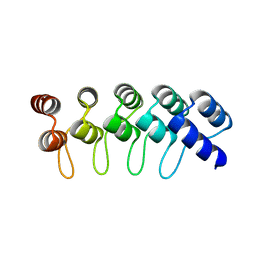 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR264. | | Descriptor: | Engineered Protein OR264 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
4HB5
 
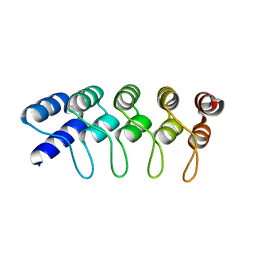 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR267. | | Descriptor: | Engineered Protein | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
5TY4
 
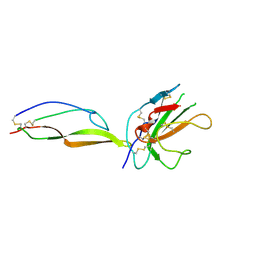 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | Descriptor: | TGF-beta receptor type-2, mmTGF-b2-7m | | Authors: | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
6ODG
 
 | | SVQIVY, Crystal Structure of a tau protein fragment | | Descriptor: | Microtubule-associated protein tau | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Seidler, P.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure-based inhibitors halt prion-like seeding by Alzheimer's disease-and tauopathy-derived brain tissue samples.
J.Biol.Chem., 294, 2019
|
|
5KWD
 
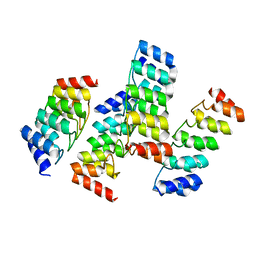 | |
4NP8
 
 | | Structure of an amyloid forming peptide VQIVYK from the second repeat region of tau (alternate polymorph) | | Descriptor: | Microtubule-associated protein tau | | Authors: | Landau, M, Eisenberg, D, Sawaya, M.R, Dannenberg, J, Kobko, N. | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular mechanisms for protein-encoded inheritance.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2PLT
 
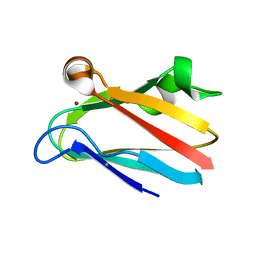 | |
2DPK
 
 | |
6TZN
 
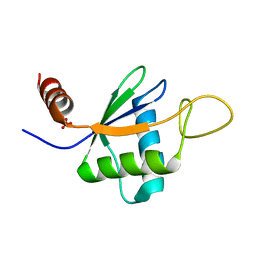 | | Structure of S. pombe telomerase accessory protein Pof8 C-terminal domain | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Basu, R.S, Cascio, D, Eichhorn, C.D, Feigon, J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of S. pombe telomerase protein Pof8 C-terminal domain is an xRRM conserved among LARP7 proteins.
Rna Biol., 18, 2021
|
|
4HXT
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR329 | | Descriptor: | De Novo Protein OR329 | | Authors: | Vorobiev, S, Su, M, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Maglaqui, M, Xiao, X, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-11-12 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
NAT.CHEM., 9, 2017
|
|
6WX1
 
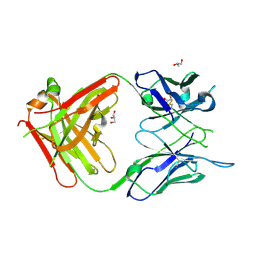 | | Antigen Binding Fragment of OKT9 | | Descriptor: | GLYCEROL, OKT9 Fab Heavy Chain, OKT9 Fab Light Chain | | Authors: | Rodriguez, J.A, Helguera, G, Sawaya, M, Cascio, D, Collazo, M, Flores, M, Zink, S, Ferrero, S, Payes, C, Fuentes, D, Short, C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antibody-Based Inhibition of Pathogenic New World Hemorrhagic Fever Mammarenaviruses by Steric Occlusion of the Human Transferrin Receptor 1 Apical Domain.
J.Virol., 95, 2021
|
|
7LUX
 
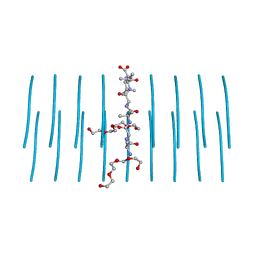 | | AALALL segment from the Nucleoprotein of SARS-CoV-2, residues 217-222, crystal form 2 | | Descriptor: | Nucleoprotein AALALL, TETRAETHYLENE GLYCOL | | Authors: | Lu, J, Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.303 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
7LTU
 
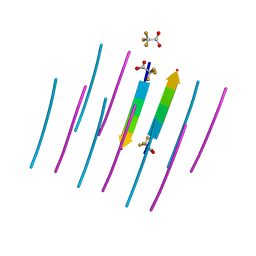 | | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2, RESIDUES 217-222, CRYSTAL FORM 1 | | Descriptor: | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2,RESIDUES 217-222, trifluoroacetic acid | | Authors: | Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.122 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
4GZR
 
 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP (Rv2346c-Rv2347c) complex in space group C2221 | | Descriptor: | ESAT-6-like protein 6, ESAT-6-like protein 7, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, He, Q, Harris, L, Zhou, T.T, Kuo, E, Ahn, C.J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-06 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4I0X
 
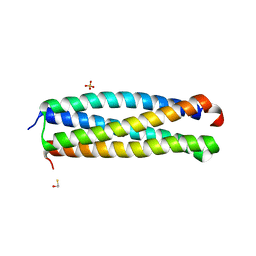 | | Crystal structure of the Mycobacterum abscessus EsxEF (Mab_3112-Mab_3113) complex | | Descriptor: | BETA-MERCAPTOETHANOL, ESAT-6-like protein MAB_3112, ESAT-6-like protein MAB_3113, ... | | Authors: | Arbing, M.A, Chan, S, Lu, J, Kuo, E, Harris, L, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-11-19 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
2LGS
 
 | | FEEDBACK INHIBITION OF FULLY UNADENYLYLATED GLUTAMINE SYNTHETASE FROM SALMONELLA TYPHIMURIUM BY GLYCINE, ALANINE, AND SERINE | | Descriptor: | GLUTAMIC ACID, GLUTAMINE SYNTHETASE, MANGANESE (II) ION | | Authors: | Liaw, S.-H, Eisenberg, D. | | Deposit date: | 1994-08-05 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Feedback inhibition of fully unadenylylated glutamine synthetase from Salmonella typhimurium by glycine, alanine, and serine.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
3Q2X
 
 | |
3PZZ
 
 | |
3NHC
 
 | | GYMLGS segment 127-132 from human prion with M129 | | Descriptor: | Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
3NHD
 
 | | GYVLGS segment 127-132 from human prion with V129 | | Descriptor: | ACETIC ACID, Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
3NVH
 
 | |
